塑料制品因其成本低、实用性高和外表美观等特点已成为人类日常生产生活中的必需品。截至2018年,世界塑料总产量已达3.59亿t,其中我国占比30%[1]。巨大的塑料产量在为人们提供便利的同时,也因对其大量消耗和不恰当处置,给自然环境和人类健康带来极大威胁。据估计,到2025年,海洋中将积累2.5亿t塑料碎片[2]。在物理、化学和生物外力的共同作用下,暴露于环境中的塑料垃圾易碎裂成小块塑料[3-4]。微塑料(microplastics, MPs)通常是指粒径<5 mm的塑料碎片[5]。其进入水环境后,由于疏水表面能快速刺激生物膜的形成,MPs为微生物的定殖和扩散提供了独特的基质和载体[6-7]。此外,由于具有比表面积大、持久性长和不易降解等特点,MPs极易吸附环境中的多氯联苯(polychlorinated biphenyls, PCBs)、多环芳烃(polycyclic aromatic hydrocarbons, PAHs)等持久性有机污染物[8]和重金属[9],从而产生一系列潜在的环境问题和生态风险。
2013年,Zettler等[10]首次在北大西洋的海洋塑料垃圾上发现了多种生物群落(包括异养生物、自养生物、捕食者和共生体),并将塑料及其附着的生物群落组合命名为“plastisphere”(塑料圈),“塑料圈”中微生物以细菌群落为主[11-12]。近年来,关于MPs上附着细菌群落多样性的研究正在兴起。MPs上附着的细菌群落主要来源于周围水体[13-15],但其群落组成和结构与周围水体和自然基质中的群落有显著差异[16-17]。MPs作为一种独特的微生物栖息地,在改变群落结构的同时,还可以潜在地影响水生生态系统中微生物群落的生态功能[18]。更加值得注意的是,Keswani等[19]的研究表明该“塑料圈”可以作为病原体、粪便指示生物和有害藻华在水环境中长期存在和传播的重要载体。此外,已有研究发现MPs表面降解菌的存在[10],并进一步证实塑料降解菌可能通过改变MPs的亲水性、表面形貌和官能团等理化性质而影响其在水环境中的迁移行为[20]。
本文从研究方法、组成、影响因素和生态效应4个方面较为全面地综述了近5年国内外水环境中MPs上细菌群落特征的研究结果,对今后开展相关工作具有指导意义,同时为MPs在水环境中归宿问题的研究提供科学依据。
1 微塑料附着生物膜研究方法进展(Advances in research methods of microplastics attached biofilm)
1.1 微塑料附着生物膜的表面形貌和立体结构
MPs在水环境中暴露较短的时间(3~24 h)即可形成细菌生物膜[14,21]。细菌生物膜是一种细菌聚集体膜状物,它是通过不可逆附着于物体表面的细菌分泌的胞外多聚物将菌体包裹其中而形成的,一般分为4个过程:细菌定殖→菌体分泌胞外多聚物→细菌增殖→形成生物膜[22]。通过观察MPs附着生物膜的表面形貌和立体结构,可从形态学角度初步判断MPs表面细菌群落的组成和结构特征。
1.1.1 生物膜的表面形貌
扫描电子显微镜(scanning electron microscope, SEM)其因样品制备方法简单、放大倍数范围广、分辨率高、成像直观、立体感强、几乎不损伤和污染原始样品等优点,可以被用来原位观察MPs附着生物膜的表面形貌特征。Arias-Andres等[23]、Wu等[17]和Shabbir等[24]使用SEM发现MPs表面附着有明显的细菌群落结构。Feng等[25]从SEM图上清晰地观察到棒状细菌和链球菌。在Guan等[26]的研究中,定殖到MPs表面的细菌主要以棒状、丝状、球形和栅栏状为主。Arias-Andres等[27]通过SEM观察到中寡营养湖泊中的MPs表面粗糙的边缘部分有斑状细菌定殖。
1.1.2 生物膜的立体结构
激光扫描共聚焦显微镜(confocal laser scanning microscope, CLSM)因其荧光成像的原理可被用于观察细胞或组织内部的细微结构[28-30],现阶段被广泛用于观察MPs附着生物膜的立体结构。Tu等[31]用荧光染色剂SYTO9、PI和Concanavalin A分别染色活细胞(染为绿色)、死细胞(染为红色)和胞外聚合物(染为蓝色),通过CLSM观察发现,MPs表面生物膜厚度随着暴露时间的延长而增加。Tarafdar等[32]用SYTO9和PI分别染色活细胞和死细胞,也获得了相似的结果。Michels等[33]和Leiser等[34]通过CLSM观察到塑料表面生物膜中的细菌和藻类细胞。Di Pippo等[35]联合荧光原位杂交技术(FISH)和CLSM技术分析评价生物膜组成和三维结构,发现细菌和微藻呈斑片状定殖在MPs表面。Schlundt等[36]开发了一种利用CLSM研究“塑料圈”群落的组合标记和光谱成像-荧光原位杂交(CLASI-FISH)方法,识别和观察了聚乙烯样品上微生物生物膜内指定分类群及其空间分布,增加了分类鉴定的可信度。
1.2 微塑料表面细菌群落结构和多样性研究
在早期研究中,以变性梯度凝胶电泳(denaturing gradient gel electrophoresis, DGGE)为代表的指纹图谱技术被广泛用于调查MPs表面的微生物群落特征,例如Oberbeckmann等[37]通过该方法对欧洲北部海域中塑料附着生物膜的微生物多样性和结构的空间和季节变化进行了研究分析。但近年来,随着分子生物学技术的快速发展,新一代高通量测序技术已被广泛应用于“塑料圈”中细菌群落组成的研究分析中[37-39]。Zettler等[10]首次运用新一代高通量测序技术,检测分析了北大西洋中塑料碎片表面微生物群落多样性及其与周围海水中群落的差异,发现塑料基质中物种均匀度更高,而平均丰富度更低。相比较DGGE,新一代高通量测序技术更加全面、准确[15]。
测序结果通常以97%的相似度聚类为操作分类单元(operational taxonomic units, OTUs),并以此为依据进行细菌群落的多样性分析。维恩图可以直观地显示出各样本间共有和特定的OTUs数量[40-41]。基于OTUs的α-多样性(Chao1、Shannon指数等)和β-多样性(主成分分析(PCA)、主坐标分析(PCoA)、非度量多维尺度分析(NMDS))分别用于反映单个样品的物种多样性和比较样本间物种多样性的相似程度[42]。利用PICRUSt等菌群代谢功能预测工具,可实现对微生物代谢功能的预测[42-44]。运用FAPROTAX软件解析微生物群落功能[44],如硝酸盐呼吸、产甲烷、发酵和植物病原等,也可用于分析海洋、湖泊中硫、碳、氢和氮的循环功能。
由此可见,MPs上细菌群落多样性的研究多以SEM和CLSM为辅助方法观察生物膜的表面形貌和立体结构,从形态学角度初步判断MPs表面细菌群落的组成和结构特征,以高通量测序技术以及测序结果的多样性分析为核心方法确定群落组成及其功能。
2 不同水域中微塑料表面的细菌群落组成(Composition of bacterial community colonizing microplastics in different aquatic environments)
2.1 海水中微塑料表面的细菌群落组成
近年来海洋作为MPs的重要“汇”,海水中MPs上细菌群落组成的研究较淡水中更为丰富,且大多数研究主要集中在近岸海水,尤其是近岸海水养殖区。已有的研究显示,海水中MPs上细菌群落的核心组成成员为变形菌、拟杆菌、蓝细菌和厚壁菌,其中优势菌群由变形菌和拟杆菌组成,如表1所示[14-15,21-22,25,31,38-40,45-52]。由表1可知,在不同海域中MPs表面的优势菌群组成略有差异。鉴于MPs表面细菌主要来源于周围环境[11,14-15],说明这种差异可能是由周围环境的微生物多样性引起[14,22]。Sun等[14]的研究表明,海水养殖区MPs表面的微生物多样性显著低于近海中的,这可能是因为海水养殖区设施数量多,造成水流减少[53],不利于不同区域之间的养分交换,最终影响了海水养殖区的细菌群落多样性[54-56];此外,Parrish和Fahrenfeld[57]的研究表明,暴露于河水中的MPs表面以变形菌为绝对优势菌门,而暴露于污水中的MPs表面,除了变形菌门外,拟杆菌门的丰度也较高,这可能由于来源于河水和污水中的MPs表面附着的浮游细菌不同。
α变形菌和γ变形菌被描述为海洋生物膜的早期定殖者[13],是海洋“塑料圈”变形菌门中最丰富的类群[22,25,38,47,51,58]。α变形菌中以红细菌科(Rhodobacteraceae)为主[15,38,45-46,58-59],且其中具有群体感应系统[60]的玫瑰杆菌分支成员是MPs表面生物膜形成和演替的普遍参与者[38]。除红细菌科外,α变形菌中的赤杆菌科(Erythrobacteraceae)和鞘脂单胞菌科(Sphingomonadaceae)也在海洋环境“塑料圈”中普遍存在且丰度较高[40,47,58-59]。此外,Bryant等[45]在北太平洋的聚乙烯和聚丙烯塑料碎片上发现生丝单胞菌科(Hyphomonadaceae)也是α变形菌中最丰富的菌科之一。γ变形菌中主要是假单胞菌科(Pseudomonadaceae)中的假单胞菌属(Pseudomonas)、假交替单胞菌科(Pseudoalteromonadaceae)中的假交替单胞菌属(Pseudoalteromonas)和交替单胞菌属(Alteromonas)[21,37,58]。Dussud等[61]在西地中海中的塑料碎片上检测到大量蓝细菌,而蓝细菌被认为是海洋塑料生物膜中的主要光自养微生物[37]。Quero和Luna[62]分析前人的研究结果得出,在近海,“塑料圈”核心微生物群包括拟杆菌门(黄杆菌科Flavobacteriaceae)和变形菌门(柄杆菌目Caulobacterales、生丝单胞菌科Hyphomonadaceae、红细菌科Rhodobacteraceae和Alcanivoracaceae)。
表1 海水中微塑料(MPs)表面细菌群落组成
Table 1 Microbial community composition on microplastics (MPs) in seawater

研究区域 Study area微塑料类型Microplastic types核心细菌群落Core bacterial community参考文献References中国山东半岛桑沟湾Sungo Bay, Shandong Peninsula, China渔网、泡沫、浮子(1~4 mm)Fishing nets, foams, floats (1~4 mm)变形菌、拟杆菌、蓝细菌、厚壁菌Proteobacteria, Bacteroidetes, Cyanobacteria, Firmicutes[14]中国东南沿海水产养殖海域Aquaculture regions of China’s southeast coastal areasPE、PP、PS、PET变形菌、拟杆菌Proteobacteria, Bacteroidetes[15]中国山东省桑沟湾海水养殖区Sungo Bay mariculture area, Shandong Province, ChinaPE、PP、PVC变形菌、拟杆菌Proteobacteria, Bacteroidetes[21]中国山东省烟台市北黄海养马岛近海养殖区Offshore aquaculture area of Northern Yellow Sea located in Yangma Island, Yantai, Shandong Province, ChinaPE变形菌、拟杆菌Proteobacteria, Bacteroidetes[22]人工/野生珊瑚区Artificial/wild coral areasPP、PC、ABS、PA、LDPE、PS、PET、EPS、PVC变形菌、拟杆菌、放线菌、Patescibacteria、厚壁菌、蓝细菌Proteobacteria, Bacteroidetes, Actinobacteria,Patescibacteria, Firmicutes, Cyanobacteria[25]中国黄海沿岸海水 Coastal seawater of Yellow Sea, ChinaPEα变形菌、γ变形菌、拟杆菌 Alphaproteobacteria, Gammaproteobacteria, Bacteroidia[31]中国南北海岸线Northern and southern Chinese coastlinesPP、PVC变形菌、拟杆菌Proteobacteria, Bacteroidetes[38]中国浙江省舟山市摘箬山岛(潮间带、潮上带、海水)Zhairuoshan Island, Zhoushan, Zhejiang Province, China (intertidal zone, supralittoral zone, seawater)PE、PET变形菌、蓝细菌、拟杆菌、放线菌Proteobacteria, Cyanobacteria, Bacteroidetes, Actinobacteria[39]中国山东省烟台市Yantai, Shandong Province, ChinaPVC变形菌、拟杆菌、蓝细菌Proteobacteria, Bacteroidetes, Cyanobacteria[40]北太平洋North PacificPE、PP蓝细菌、α变形菌、拟杆菌Cyanobacteria, Alphaproteobacteria, Bacteroidetes[45]北大西洋North AtlanticPEβ变形菌Betaproteobacteria[46]北亚得里亚海North Adriatic SeaPE、PP变形菌、浮霉菌、绿弯菌、拟杆菌、蓝细菌、厚壁菌Proteobacteria, Planctomycetes, Chloroflexi, Bacteroidetes, Cyanobacteria, Firmicutes[47]波罗的海The BalticPE、PP、PSα变形菌、β变形菌、拟杆菌、浮霉菌Alphaproteobacteria, Betaproteobacteria, Bacteroidetes, Planctomycetes[48]布雷斯特湾 The Bay of BrestPE、PP、PSα变形菌、γ变形菌、拟杆菌、蓝细菌Alphaproteobacteria, Gammaproteobacteria, Bacteroidetes, Cyanobacteria[49]新加坡海岸线Coastlines of Singapore泡沫颗粒(55%)和碎片(35%)Foam particles (55%) and fragments (35%)变形菌、拟杆菌Proteobacteria, Bacteroidetes[50]
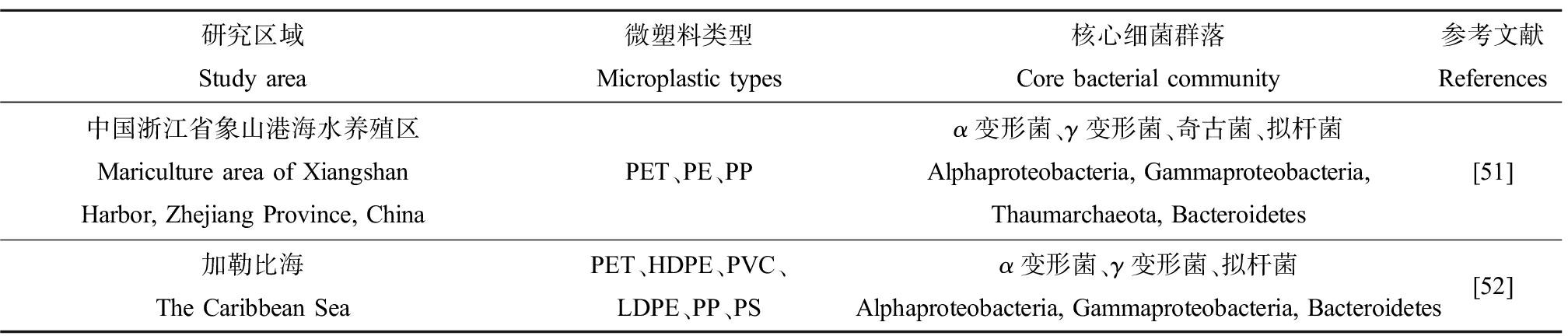
研究区域 Study area微塑料类型Microplastic types核心细菌群落Core bacterial community参考文献References中国浙江省象山港海水养殖区Mariculture area of XiangshanHarbor, Zhejiang Province, ChinaPET、PE、PPα变形菌、γ变形菌、奇古菌、拟杆菌Alphaproteobacteria, Gammaproteobacteria, Thaumarchaeota, Bacteroidetes[51]加勒比海 The Caribbean SeaPET、HDPE、PVC、LDPE、PP、PSα变形菌、γ变形菌、拟杆菌Alphaproteobacteria, Gammaproteobacteria, Bacteroidetes[52]
注:PE为聚乙烯;PET为聚对苯二甲酸乙二醇酯;PP为聚丙烯;PVC为聚氯乙烯;PA为尼龙;PS为聚苯乙烯;LDPE为低密度聚乙烯;EPS为发泡聚苯乙烯;ABS为丙烯腈、丁二烯、苯乙烯3种单体的三元共聚物;PC为聚碳酸酯;HDPE为高密度聚乙烯。
Note: PE is polyethylene; PET is polyethylene terephthalate; PP is polypropylene; PVC is polyvinyl chloride; PA is nylon; PS is polystyrene; LDPE is low density polyethylene; EPS is expanded polystyrene; ABS is acrylonitrile butadiene styrene plastic; PC is polycarbonate; HDPE is high density polyethylene.
2.2 淡水中微塑料表面的细菌群落组成
淡水环境中MPs上细菌群落组成的研究主要集中在城市水环境中,且与海水环境中MPs表面的细菌群落有相似之处,即变形菌、拟杆菌为优势菌群,除此之外,蓝细菌和厚壁菌也是核心群落的成员(表2)[17-18,24,26-27,35,42,63-65]。
类似于海水中的研究,变形菌被认为是淡水MPs生物膜上早期定殖体的常见组成成员[63],主要由α变形菌(红细菌科Rhodobacteraceae、鞘脂单胞菌科Sphingomonadaceae)和γ变形菌(假单胞菌科Pseudomonadaceae)为代表[16,35,63-64,66];拟杆菌中主要包括Sphingosinobacteriaceae和黄杆菌科(Flavobacteriaceae)[16,35,63]。此外,丛毛单胞菌科(Comamonadaceae)[12,63]、气单胞菌科(Aeromonadaceae)[12,16,63]、莫拉菌科(Moraxellaceae)[12,63]和弯曲杆菌科(Campylobacteraceae)[12,16]也被发现是淡水MPs生物膜中的丰富科。
综上,变形菌(α变形菌、γ变形菌)和拟杆菌是水环境“塑料圈”中的优势菌群,蓝细菌和厚壁菌也是水环境“塑料圈”中核心群落的组成成员,放线菌次之,而海水相较于淡水,浮霉菌也可能作为核心群落组成成员。此外,徐沛等[67]研究发现,由于人类活动的过度干扰,MPs存在于长江口及其邻近海域的表面沉积物中已是一种普遍现象,但关于河口系统中MPs表面细菌群落组成的研究仍较少。Jiang等[43]通过高通量测序技术分析了长江口潮间带MPs样品的细菌群落组成,发现核心群落主要包括变形菌门、蓝藻门、拟杆菌门和放线菌门,并用相关网络分析方法鉴定了重要的细菌种类(红细菌目Rhodobacterales、鞘脂单胞菌目Sphingomonadales和根瘤菌目Rhizobiales)。Baptista Neto等[68]在维多利亚湾河口系统沉积物中的渔网碎片微纤维表面检测到的微生物最主要是细菌,其次是硅藻残余物、真菌丝和孢子。
3 影响微塑料表面细菌群落多样性的因素(Factors influencing diversity of bacterial community colonizing microplastics)
3.1 微塑料暴露时间对其附着细菌群落多样性的影响
Xu等[38,69]和Li等[39]的研究表明在海洋生境中,随着MPs暴露时间的延长,生物膜上细菌群落多样性呈现逐渐增加的趋势。细菌群落在MPs生物膜上的生长是一个时间演替过程,可分为早期、中期和晚期定殖[70],生物膜形成阶段也是海洋塑料垃圾上细菌群落多样性的影响因素之一[61]。De Tender等[11]的研究表明细菌群落在塑料碎片上的定殖存在时间梯度。随着暴露时间的推移,α变形菌、β变形菌和黄杆菌的相对丰度增加,γ变形菌的相对丰度减少。此外,弧菌是海洋环境中聚乙烯和聚苯乙烯MPs的早期定殖体[58],而在定殖后期阶段黄杆菌科(Flavobacteriaceae)、红细菌科(Rhodobacteraceae)、浮霉菌科(Planctomycetaceae)和叶杆菌科(Phyllobacteriaceae)的数量较多[71]。
3.2 微塑料附着细菌群落多样性的空间差异性
已有研究证明海岸、河口和河流中MPs表面附着的细菌群落组成和多样性均会因监测点的不同而显著不同[43,50,72],且徐希媛[69]的研究表明,暴露海域对生物膜中细菌群落的多样性影响最大,暴露时间其次。“塑料圈”细菌群落多样性的空间差异主要是由盐度、温度、酸碱度和有机物等环境因素以及周围环境微生物群落特征的不同引起的[13-14,38,73],且Wang等[42]、Kesy等[58]和Li等[74]的研究结果显示盐度是主要因素。MPs附着细菌群落特征随暴露深度的变化通常伴随着周围水温、光强、pH和溶解氧等环境因素以及水体中微生物群落特征的变化,这可能是影响MPs表面生物膜的微生物群落结构与多样性的原因[11,22]。
3.3 微塑料理化性质对其附着细菌群落多样性的影响
关于MPs类型是否会影响生物膜中微生物群落多样性,前人的研究结果并不一致。Di Pippo等[35]、Xu等[38]、Li等[39]和Wu等[41]的研究表明MPs类型对其附着微生物群落的组成、结构和丰度没有显著影响。而Feng等[25]、Parrish和Fahrenfeld[57]的研究表明MPs类型是影响其生物膜上微生物群落结构、丰度的重要因素。据此,前人对MPs理化性质方面进一步分析。Cai等[75]的研究表明细菌附着在不同类型的塑料上可以由塑料的物理化学性质来控制,并发现塑料表面硬度是影响细菌在塑料表面定殖的关键因素。此外,塑料表面可用性[51]、粗糙度[25,63,76]、疏水性[63,76]以及颗粒大小[51]也可能影响细菌在塑料表面的定殖过程,从而影响表面细菌群落多样性。除了MPs表面理化性质以外,柴然[21]和Xie等[77]的研究表明细菌在MPs上的选择性定殖可能是由于细菌对聚合物中某些化学物的趋化性所导致。被MPs吸附的污染物[65,78]、塑料中的添加剂[13]以及外源化合物[24]可能也会选择性地促进或抑制不同细菌物种的生长,影响生物膜上的细菌群落的多样性。
表2 淡水中微塑料(MPs)表面细菌群落组成
Table 2 Microbial community composition on microplastics (MPs) in freshwater
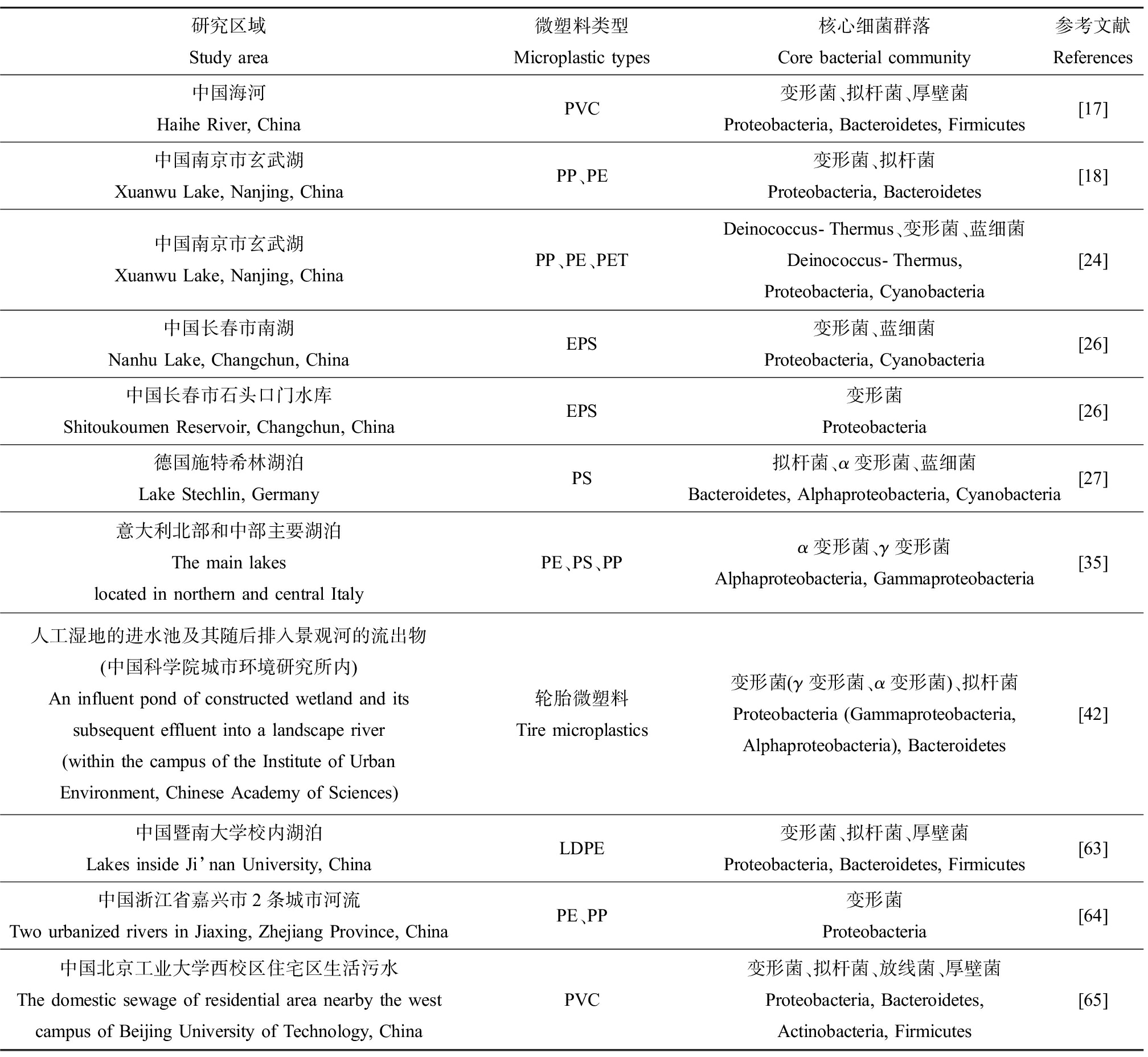
研究区域Study area微塑料类型Microplastic types核心细菌群落Core bacterial community参考文献References中国海河Haihe River, ChinaPVC变形菌、拟杆菌、厚壁菌Proteobacteria, Bacteroidetes, Firmicutes[17]中国南京市玄武湖Xuanwu Lake, Nanjing, ChinaPP、PE变形菌、拟杆菌Proteobacteria, Bacteroidetes[18]中国南京市玄武湖Xuanwu Lake, Nanjing, ChinaPP、PE、PETDeinococcus-Thermus、变形菌、蓝细菌Deinococcus-Thermus, Proteobacteria, Cyanobacteria[24]中国长春市南湖Nanhu Lake, Changchun, ChinaEPS变形菌、蓝细菌Proteobacteria, Cyanobacteria[26]中国长春市石头口门水库Shitoukoumen Reservoir, Changchun, ChinaEPS变形菌Proteobacteria[26]德国施特希林湖泊Lake Stechlin, GermanyPS拟杆菌、α变形菌、蓝细菌Bacteroidetes, Alphaproteobacteria, Cyanobacteria[27]意大利北部和中部主要湖泊The main lakeslocated in northern and central ItalyPE、PS、PPα变形菌、γ变形菌Alphaproteobacteria, Gammaproteobacteria[35]人工湿地的进水池及其随后排入景观河的流出物(中国科学院城市环境研究所内)An influent pond of constructed wetland and its subsequent effluent into a landscape river (within the campus of the Institute of Urban Environment, Chinese Academy of Sciences)轮胎微塑料Tire microplastics变形菌(γ变形菌、α变形菌)、拟杆菌Proteobacteria (Gammaproteobacteria, Alphaproteobacteria), Bacteroidetes[42]中国暨南大学校内湖泊Lakes inside Ji’nan University, ChinaLDPE变形菌、拟杆菌、厚壁菌Proteobacteria, Bacteroidetes, Firmicutes[63]中国浙江省嘉兴市2条城市河流Two urbanized rivers in Jiaxing, Zhejiang Province, ChinaPE、PP变形菌Proteobacteria[64]中国北京工业大学西校区住宅区生活污水The domestic sewage of residential area nearby the west campus of Beijing University of Technology, ChinaPVC变形菌、拟杆菌、放线菌、厚壁菌Proteobacteria, Bacteroidetes, Actinobacteria, Firmicutes[65]
综上,暴露时间、空间以及自身理化性质都可能是影响MPs表面附着细菌群落多样性的重要因素。事实上,水环境中MPs表面的细菌群落多样性并不是单一影响因素作用的结果,而是由多种因素相互作用产生的[11],但在特定情况下可能会由某种影响因素发挥主导作用,例如Oberbeckmann等[59]研究发现,总体上MPs表面参数对附着生物膜细菌群落的影响次于环境因素,但在细菌定殖MPs的早期阶段,表面参数起主导作用,且空间因素的重要性随着时间的推移而增加;而另一种解释为相较于表面参数,环境因素可能通过影响调节膜的形成从早期定殖阶段起就对细菌群落在MPs表面的定殖具有更大的影响。
4 微塑料上细菌群落的生态效应(Ecological effects of bacterial community on microplastics)
4.1 微塑料作为潜在病原体的载体
已有研究发现潜在病原体是塑料附着微生物群落的“搭便车者”[43]。Viršek等[47]在亚得里亚海北部MPs上首次鉴定出致病性鱼类细菌杀鲑气单胞菌。Gong等[63]的研究表明,我国暨南大学一个校园湖泊中在低密度聚乙烯上定居的丰度前20位的细菌属中有一半是潜在的病原体,例如植物病原体土壤杆菌、医院病原体金黄杆菌和鱼类病原体黄杆菌。
弧菌作为海洋中一种普遍存在的潜在致病菌,前人的研究已证实其在MPs上普遍存在[14,38-39],弧菌属的许多物种对人类以及鱼、虾和贝类等海洋动物具有致病作用,同时弧菌也是珊瑚白化、溶解、白带和黄斑病的病原体[25]。例如,Kirstein等[79]在来自北海/波罗的海的聚乙烯、聚丙烯和聚苯乙烯MPs颗粒上检测到了潜在致病性副溶血弧菌;柴然[21]在暴露于养殖海域的MPs上检测到了属于弧菌属的海洋生物病原菌,如副溶血弧菌和创伤弧菌等。除弧菌属外,假单胞菌属也是水环境中MPs上常见的潜在致病菌属[12,15,25,39,41,43,50]。更加值得注意的是,已有较多的研究结果表明,与周围水体和自然基质相比,MPs表面定殖的弧菌、假单胞菌等潜在致病菌的相对丰度更高[14,21,25,41,58]。此外,Kesy等[58]和Li等[74]发现MPs上弧菌属的丰度和水环境中盐度呈正相关;江沛霖[15]研究发现夏季MPs样品中潜在病原菌的丰度占比相较于冬季更高。由此可见,环境因素是影响水环境中MPs表面生物膜中潜在病原体丰度的重要原因。
4.2 微塑料表面存在塑料降解菌
Xu等[38]、Li等[39]和Dussud等[61]通过SEM观察到了MPs表面的降解迹象;Shabbir等[24]在质量损失估算的基础上,利用SEM、红外光谱和凝胶渗透色谱观察到的MPs表面的形态变化进一步证实了生物降解;Niu等[20]的研究表明,塑料降解菌是MPs降解的关键因素,并通过塑料降解菌在MPs上的丰度和介数中心性的增加,以及使用接触角测量、SEM和傅里叶变换红外光谱对MPs进行表征,证实了较深的沉积层可能促进MPs的生物降解。McCormick等[12,72]的研究表明假单胞菌科在MPs表面有富集现象,假单胞菌是降解高密度聚乙烯[80]、低密度聚乙烯[81]和聚丙烯[82]等塑料聚合物的相关细菌;且假单胞菌属能够产生丝氨酸水解酶、酯酶和脂肪酶等酶系,这些酶有助于塑料的生物降解[83]。除假单胞菌以外,已有研究还发现红杆菌属[50]、芽孢杆菌属[63]和黄杆菌属[35]等烃降解菌是MPs表面生物膜中的优势菌。Zettler等[10]在北大西洋的研究中通过SEM和小亚基rRNA基因鉴定出了几种仅存在于塑料碎片表面而在海水中没有检测到的烃降解细菌;此外,他们通过网络分析发现,虽然潜在烃降解分类群的单独存在并不意味着它们与塑料降解有关,但OTUs的联合体可能共同作用来利用顽固的碳源,这对于未来塑料降解菌的研究可能具有重要意义。
4.3 微塑料表面微生物携带并传播抗生素抗性基因
已有研究表明水环境包括污水中的MPs微生物对抗生素抗性基因(antibiotic resistance genes, ARGs)具有富集作用[17,64-65]。Yang等[84]通过分析北太平洋环流塑料颗粒的宏基因组数据,发现多药抗性基因和多金属抗性基因是塑料微生物群中检测到的主要基因类别,并首次报道总ARGs(64个亚型)在MPs中的相对丰度显著大于海水(P<0.05);Wu等[17]研究发现MPs生物膜中相对丰度最高的ARGs亚型属于多药抗性型,MPs生物膜可作为金属和ARGs的共同选择和转移的焦点[85]。进一步的研究表明水环境中MPs可能是水平基因转移的热点[59],相较于水中的浮游细菌群落,MPs中的水平基因转移可能具有更高的水平[64],例如ARGs。值得注意的是MPs生物膜中ARGs的微生物宿主,例如Wu等[17]在MPs上检测到的2种机会性人类病原体(Pseudomonas monteilii、Pseudomonas mendocina)和1种植物病原体(Pseudomonas syringae)。综上来看,MPs可能作为抗性基因进入新环境的载体,对水生生态系统产生潜在风险并对人类健康产生不利影响。

图1 水环境中微塑料的附着生物膜
注:ARG为抗生素抗性基因;MRG为金属抗性基因;EPS表示胞外聚合物。
Fig. 1 Microplastic-associated biofilm in the aquatic environment
Note: ARG stands for antibiotic resistance gene; MRG stands for metal resistance gene; EPS stands for extracellular polymeric substances.
4.4 微塑料附着细菌群落影响水环境中的生物地球化学循环
鉴于MPs表面的微生物群落多样性不同于周围水体[16,72],作为一种独特的微生物栖息地,其附着的微生物群落具有不同于自由生活的浮游微生物群落的生活方式、代谢途径和生物地球化学活动[45],从而可能影响水环境中的生物地球化学循环。MPs表面的群体感应细菌[43]和其他富集的细菌物种[14](弧菌、假交替单胞菌和交替单胞菌)分别在MPs周围碳循环和碳代谢中起到重要作用,因此未来应进一步研究MPs附着细菌对与碳水化合物代谢相关的海洋碳循环的影响。Dussud等[61]在西地中海中的塑料碎片上检测到大量蓝细菌,并指出它们可能影响地表海洋的全球碳、氮生物地球化学循环。Chen等[86]的研究表明,MPs表面的附着生物膜可能通过提高反硝化能力影响淡水中的氮循环,通过吸附和微生物介导的磷转化影响淡水中的磷循环。Xie等[77]在暴露于红树林生态系统中的MPs附着生物膜上检测到了对于碳循环或硫循环具有重要意义的独特优势菌群。
综上来看,MPs一方面为附着细菌群落及其可移动遗传元件在水环境中的远距离运输、分散提供了载体,这些可移动遗传元件包括ARGs和致病菌[27,87](图1),从而可能危害生态系统安全和人类健康。另一方面,“塑料圈”上细菌群落的多样性影响水环境中的生物地球化学循环。此外,由于MPs上塑料降解菌的发现,增加了塑料降解菌筛选技术的可行性。
5 结论与展望(Conclusions and perspectives)
综上,笔者对本领域研究现状总结如下:
(1)MPs上细菌群落多样性的研究以高通量测序技术以及测序结果的多样性分析为核心方法,以SEM和CLSM为辅助方法。
(2)海水、淡水环境中MPs表面附着的细菌群落均以变形菌门、拟杆菌门占主导。
(3)暴露时间、暴露地点以及自身理化性质均可能是影响MPs表面附着细菌群落多样性的重要因素。
(4)MPs可以作为ARGs、致病菌和降解菌在水环境中迁移的载体,且MPs表面的细菌群落影响水环境中的生物地球化学循环。
目前MPs附着生物膜的研究主要集中在细菌群落,对“塑料圈”中的藻类、真菌等其他生物的研究较为匮乏,亟待更多的基础研究;水环境研究范围分布不均衡,主要集中在近海环境,因此应加强远洋、入海口和内陆地表水,尤其是城市水环境中的研究,以此探索全球范围水环境中MPs表面微生物群落的定殖规律及其生态效应。此外,虽然已有较多研究指出MPs表面附着生物膜具有潜在生态风险,但关于MPs及其附着生物膜对水体/沉积环境、水生生物和人类的联合毒性及其作用机制亟待进一步揭示。在继续加强基础信息积累的同时,未来的研究方向应更加侧重于MPs污染防治的对策和措施的制定、生态风险评价等方面。
通讯作者简介:邓惠(1988—),女,博士后,主要研究方向为新型污染物环境行为。
共同通讯作者简介:葛成军(1977—),男,博士,教授,主要研究方向为污染物环境行为与健康风险。
[1] 许江菱. 2019—2020年世界塑料工业进展(Ⅰ): 通用塑料[J]. 塑料工业, 2021, 49(3): 1-9
Xu J L. Progress of the world’s plastics industry in 2019-2020 (Ⅰ): General purposed plastics [J]. China Plastics Industry, 2021, 49(3): 1-9 (in Chinese)
[2] Jambeck J R, Geyer R, Wilcox C, et al. Marine pollution. Plastic waste inputs from land into the ocean [J]. Science, 2015, 347(6223): 768-771
[3] Andrady A L. Microplastics in the marine environment [J]. Marine Pollution Bulletin, 2011, 62(8): 1596-1605
[4] Anderson J C, Park B J, Palace V P. Microplastics in aquatic environments: Implications for Canadian ecosystems [J]. Environmental Pollution, 2016, 218: 269-280
[5] Thompson R C, Olsen Y, Mitchell R P, et al. Lost at sea: Where is all the plastic? [J]. Science, 2004, 304(5672): 838
[6] Oberbeckmann S, Löder M G J, Labrenz M. Marine microplastic-associated biofilms: A review [J]. Environmental Chemistry, 2015, 12(5): 551
[7] Amaral-Zettler L A, Zettler E R, Mincer T J. Ecology of the plastisphere [J]. Nature Reviews Microbiology, 2020, 18(3): 139-151
[8] Hirai H, Takada H, Ogata Y, et al. Organic micropollutants in marine plastics debris from the open ocean and remote and urban beaches [J]. Marine Pollution Bulletin, 2011, 62(8): 1683-1692
[9] Selvam S, Jesuraja K, Venkatramanan S, et al. Hazardous microplastic characteristics and its role as a vector of heavy metal in groundwater and surface water of coastal south India [J]. Journal of Hazardous Materials, 2021, 402: 123786
[10] Zettler E R, Mincer T J, Amaral-Zettler L A. Life in the “plastisphere”: Microbial communities on plastic marine debris [J]. Environmental Science & Technology, 2013, 47(13): 7137-7146
[11] De Tender C, Devriese L I, Haegeman A, et al. Temporal dynamics of bacterial and fungal colonization on plastic debris in the North Sea [J]. Environmental Science & Technology, 2017, 51(13): 7350-7360
[12] McCormick A, Hoellein T J, Mason S A, et al. Microplastic is an abundant and distinct microbial habitat in an urban river [J]. Environmental Science & Technology, 2014, 48(20): 11863-11871
[13] de Tender C A, Devriese L I, Haegeman A, et al. Bacterial community profiling of plastic litter in the Belgian part of the North Sea [J]. Environmental Science & Technology, 2015, 49(16): 9629-9638
[14] Sun X M, Chen B J, Xia B, et al. Impact of mariculture-derived microplastics on bacterial biofilm formation and their potential threat to mariculture: A case in situ study on the Sungo Bay, China [J]. Environmental Pollution, 2020, 262: 114336
[15] 江沛霖. 中国东南沿海部分区域微塑料附着微生物研究[D]. 上海: 华东师范大学, 2018: 32-79
Jiang P L. Microplastic-associated bacterial assemblages in some coastal areas of southeast China[D]. Shanghai: East China Normal University, 2018: 32-79 (in Chinese)
[16] Hoellein T J, McCormick A R, Hittie J, et al. Longitudinal patterns of microplastic concentration and bacterial assemblages in surface and benthic habitats of an urban river [J]. Freshwater Science, 2017, 36(3): 491-507
[17] Wu X J, Pan J, Li M, et al. Selective enrichment of bacterial pathogens by microplastic biofilm [J]. Water Research, 2019, 165: 114979
[18] Miao L Z, Wang P F, Hou J, et al. Distinct community structure and microbial functions of biofilms colonizing microplastics [J]. The Science of the Total Environment, 2019, 650(Pt 2): 2395-2402
[19] Keswani A, Oliver D M, Gutierrez T, et al. Microbial hitchhikers on marine plastic debris: Human exposure risks at bathing waters and beach environments [J]. Marine Environmental Research, 2016, 118: 10-19
[20] Niu L H, Li Y Y, Li Y, et al. New insights into the vertical distribution and microbial degradation of microplastics in urban river sediments [J]. Water Research, 2021, 188: 116449
[21] 柴然. 桑沟湾养殖区微塑料附着细菌的群落结构特征研究[D]. 青岛: 青岛大学, 2020: 3-17
Chai R. Community structure characteristics of bacteria attached on microplastics in Sanggou Bay mariculture area[D]. Qingdao: Qingdao University, 2020: 3-17 (in Chinese)
[22] 陈涛. 近海微塑料表面生物膜的形成及其对微塑料理化性质的影响[D]. 烟台: 中国科学院大学, 2018: 19-67
Chen T. Formation of biofilm on microplastics and its influences on physicochemical properties of microplastics in the coastal sea[D]. Yantai: University of Chinese Academy of Sciences, 2018: 19-67 (in Chinese)
[23] Arias-Andres M, Kettner M T, Miki T, et al. Microplastics: New substrates for heterotrophic activity contribute to altering organic matter cycles in aquatic ecosystems [J]. Science of the Total Environment, 2018, 635: 1152-1159
[24] Shabbir S, Faheem M, Ali N, et al. Periphytic biofilm: An innovative approach for biodegradation of microplastics [J]. Science of the Total Environment, 2020, 717: 137064
[25] Feng L M, He L, Jiang S Q, et al. Investigating the composition and distribution of microplastics surface biofilms in coral areas [J]. Chemosphere, 2020, 252: 126565
[26] Guan J N, Qi K, Wang J Y, et al. Microplastics as an emerging anthropogenic vector of trace metals in freshwater: Significance of biofilms and comparison with natural substrates [J]. Water Research, 2020, 184: 116205
[27] Arias-Andres M, Klümper U, Rojas-Jimenez K, et al. Microplastic pollution increases gene exchange in aquatic ecosystems [J]. Environmental Pollution, 2018, 237: 253-261
[28] 刘云. 双光子荧光显微镜扫描控制与成像系统研究[D]. 北京: 中国科学院大学, 2015: 5
Liu Y. Study of scanning control and imaging system of two photon fluorescence microscopy[D]. Beijing: University of Chinese Academy of Sciences, 2015: 5 (in Chinese)
[29] 郝立凯, 郭圆, 江娜, 等. 激光扫描共聚焦荧光显微镜技术及其在地球生物学中的应用[J]. 矿物岩石地球化学通报, 2020, 39(6): 1141-1172, 1065
Hao L K, Guo Y, Jiang N, et al. Confocal laser scanning microscopy and its application in geobiology[J]. Bulletin of Mineralogy, Petrology and Geochemistry, 2020, 39(6): 1141-1172, 1065 (in Chinese)
[30] Brakenhoff G J. 激光共焦显微成像的最新进展及其在生命科学研究中的应用[J]. 生命科学, 2009, 21(2): 191-197
Brakenhoff G J. Development of confocal laser scanning microscope technology in biomedical application[J]. Chinese Bulletin of Life Sciences, 2009, 21(2): 191-197 (in Chinese)
[31] Tu C, Chen T, Zhou Q, et al. Biofilm formation and its influences on the properties of microplastics as affected by exposure time and depth in the seawater [J]. The Science of the Total Environment, 2020, 734: 139237
[32] Tarafdar A, Lee J U, Jeong J E, et al. Biofilm development of Bacillus siamensis ATKU1 on pristine short chain low-density polyethylene: A case study on microbe-microplastics interaction [J]. Journal of Hazardous Materials, 2021, 409: 124516
[33] Michels J, Stippkugel A, Lenz M, et al. Rapid aggregation of biofilm-covered microplastics with marine biogenic particles [J]. Proceedings Biological Sciences, 2018, 285(1885): 20181203
[34] Leiser R, Jongsma R, Bakenhus I, et al. Interaction of cyanobacteria with calcium facilitates the sedimentation of microplastics in a eutrophic reservoir [J]. Water Research, 2021, 189: 116582
[35] Di Pippo F, Venezia C, Sighicelli M, et al. Microplastic-associated biofilms in lentic Italian ecosystems [J]. Water Research, 2020, 187: 116429
[36] Schlundt C, Mark Welch J L, Knochel A M, et al. Spatial structure in the “Plastisphere”: Molecular resources for imaging microscopic communities on plastic marine debris [J]. Molecular Ecology Resources, 2020, 20(3): 620-634
[37] Oberbeckmann S, Loeder M G, Gerdts G, et al. Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in Northern European waters [J]. FEMS Microbiology Ecology, 2014, 90(2): 478-492
[38] Xu X Y, Wang S, Gao F L, et al. Marine microplastic-associated bacterial community succession in response to geography, exposure time, and plastic type in China’s coastal seawaters [J]. Marine Pollution Bulletin, 2019, 145: 278-286
[39] Li J J, Huang W, Jiang R J, et al. Are bacterial communities associated with microplastics influenced by marine habitats? [J]. The Science of the Total Environment, 2020, 733: 139400
[40] Wang J H, Lu J, Zhang Y X, et al. Unique bacterial community of the biofilm on microplastics in coastal water [J]. Bulletin of Environmental Contamination and Toxicology, 2021, 107(4): 597-601
[41] Wu N, Zhang Y, Zhao Z, et al. Colonization characteristics of bacterial communities on microplastics compared with ambient environments (water and sediment) in Haihe Estuary [J]. The Science of the Total Environment, 2020, 708: 134876
[42] Wang L Y, Luo Z X, Zhen Z, et al. Bacterial community colonization on tire microplastics in typical urban water environments and associated impacting factors [J]. Environmental Pollution, 2020, 265(Pt B): 114922
[43] Jiang P L, Zhao S Y, Zhu L X, et al. Microplastic-associated bacterial assemblages in the intertidal zone of the Yangtze Estuary [J]. The Science of the Total Environment, 2018, 624: 48-54
[44] Li C C, Gan Y D, Dong J Y, et al. Impact of microplastics on microbial community in sediments of the Huangjinxia Reservoir-water source of a water diversion project in Western China [J]. Chemosphere, 2020, 253: 126740
[45] Bryant J A, Clemente T M, Viviani D A, et al. Diversity and activity of communities inhabiting plastic debris in the North Pacific Gyre [J]. mSystems, 2016, 1(3): e00024-e00016
[46] Debroas D, Mone A, Ter Halle A. Plastics in the North Atlantic garbage patch: A boat-microbe for hitchhikers and plastic degraders [J]. Science of the Total Environment, 2017, 599-600: 1222-1232
[47] Viršek M K, Lovšin M N, Koren  , et al. Microplastics as a vector for the transport of the bacterial fish pathogen species Aeromonas salmonicida [J]. Marine Pollution Bulletin, 2017, 125(1-2): 301-309
, et al. Microplastics as a vector for the transport of the bacterial fish pathogen species Aeromonas salmonicida [J]. Marine Pollution Bulletin, 2017, 125(1-2): 301-309
[48] Ogonowski M, Motiei A, Ininbergs K, et al. Evidence for selective bacterial community structuring on microplastics [J]. Environmental Microbiology, 2018, 20(8): 2796-2808
[49] Frère L, Maignien L, Chalopin M, et al. Microplastic bacterial communities in the Bay of Brest: Influence of polymer type and size [J]. Environmental Pollution, 2018, 242: 614-625
[50] Curren E, Leong S C Y. Profiles of bacterial assemblages from microplastics of tropical coastal environments [J]. Science of the Total Environment, 2019, 655: 313-320
[51] Hou D D, Hong M, Wang K, et al. Prokaryotic community succession and assembly on different types of microplastics in a mariculture cage [J]. Environmental Pollution, 2021, 268(Pt A): 115756
[52] Dudek K L, Cruz B N, Polidoro B, et al. Microbial colonization of microplastics in the Caribbean Sea [J]. Limnology and Oceanography Letters, 2020, 5(1): 5-17
[53] Shi J, Wei H, Zhao L, et al. A physical-biological coupled aquaculture model for a suspended aquaculture area of China [J]. Aquaculture, 2011, 318(3-4): 412-424
[54] Chen C S, Ji R B, Zheng L Y, et al. Influences of physical processes on the ecosystem in Jiaozhou Bay: A coupled physical and biological model experiment [J]. Journal of Geophysical Research: Oceans, 1999, 104(C12): 29925-29949
[55] Fiore C L, Jarett J K, Olson N D, et al. Nitrogen fixation and nitrogen transformations in marine symbioses [J]. Trends in Microbiology, 2010, 18(10): 455-463
[56] Sjöstedt J, Koch-Schmidt P, Pontarp M, et al. Recruitment of members from the rare biosphere of marine bacterioplankton communities after an environmental disturbance [J]. Applied and Environmental Microbiology, 2012, 78(5): 1361-1369
[57] Parrish K, Fahrenfeld N L. Microplastic biofilm in fresh- and wastewater as a function of microparticle type and size class [J]. Environmental Science: Water Research & Technology, 2019, 5(3): 495-505
[58] Kesy K, Oberbeckmann S, Kreikemeyer B, et al. Spatial environmental heterogeneity determines young biofilm assemblages on microplastics in Baltic Sea mesocosms [J]. Frontiers in Microbiology, 2019, 10: 1665
[59] Oberbeckmann S, Kreikemeyer B, Labrenz M. Environmental factors support the formation of specific bacterial assemblages on microplastics [J]. Frontiers in Microbiology, 2017, 8: 2709
[60] McDougald D, Rice S A, Kjelleberg S. Bacterial quorum sensing and interference by naturally occurring biomimics [J]. Analytical and Bioanalytical Chemistry, 2007, 387(2): 445-453
[61] Dussud C, Meistertzheim A L, Conan P, et al. Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters [J]. Environmental Pollution, 2018, 236: 807-816
[62] Quero G M, Luna G M. Surfing and dining on the “plastisphere”: Microbial life on plastic marine debris [J]. Advances in Oceanography and Limnology, 2017, 8(2): 199-207
[63] Gong M T, Yang G Q, Zhuang L, et al. Microbial biofilm formation and community structure on low-density polyethylene microparticles in lake water microcosms [J]. Environmental Pollution, 2019, 252(Pt A): 94-102
[64] Wang J, Qin X, Guo J B, et al. Evidence of selective enrichment of bacterial assemblages and antibiotic resistant genes by microplastics in urban rivers [J]. Water Research, 2020, 183: 116113
[65] Zhao Y F, Gao J F, Wang Z Q, et al. Responses of bacterial communities and resistance genes on microplastics to antibiotics and heavy metals in sewage environment [J]. Journal of Hazardous Materials, 2021, 402: 123550
[66] Qi K, Lu N, Zhang S Q, et al. Uptake of Pb(Ⅱ) onto microplastic-associated biofilms in freshwater: Adsorption and combined toxicity in comparison to natural solid substrates [J]. Journal of Hazardous Materials, 2021, 411: 125115
[67] 徐沛, 彭谷雨, 朱礼鑫, 等. 长江口微塑料时空分布及风险评价[J]. 中国环境科学, 2019, 39(5): 2071-2077
Xu P, Peng G Y, Zhu L X, et al. Spatial-temporal distribution and pollution load of microplastics in the Changjiang Estuary[J]. China Environmental Science, 2019, 39(5): 2071-2077 (in Chinese)
[68] Baptista Neto J A, Gaylarde C, Beech I, et al. Microplastics and attached microorganisms in sediments of the Vitória Bay estuarine system in SE Brazil [J]. Ocean & Coastal Management, 2019, 169: 247-253
[69] 徐希媛. 中国近海暴露环境下微塑料生物膜中生物群落时空变化及群体感应细菌研究[D]. 青岛: 自然资源部第一海洋研究所, 2020: 4-45
[70] Yang Y Y, Liu W Z, Zhang Z L, et al. Microplastics provide new microbial niches in aquatic environments [J]. Applied Microbiology and Biotechnology, 2020, 104(15): 6501-6511
[71] Pinto M, Langer T M, Hüffer T, et al. The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession [J]. PLoS One, 2019, 14(6): e0217165
[72] McCormick A R, Hoellein T J, London M G, et al. Microplastic in surface waters of urban rivers: Concentration, sources, and associated bacterial assemblages [J]. Ecosphere, 2016, 7(11): e01556
[73] 龚梦婷. 水体环境中微塑料表面的微生物群落结构分析[D]. 广州: 暨南大学, 2019: 3
Gong M T. Microbial community structure of microplastics biofilm in aquatic environments[D]. Guangzhou: Jinan University, 2019: 3 (in Chinese)
[74] Li W J, Zhang Y, Wu N, et al. Colonization characteristics of bacterial communities on plastic debris influenced by environmental factors and polymer types in the Haihe Estuary of Bohai Bay, China [J]. Environmental Science & Technology, 2019, 53(18): 10763-10773
[75] Cai L, Wu D, Xia J H, et al. Influence of physicochemical surface properties on the adhesion of bacteria onto four types of plastics [J]. Science of the Total Environment, 2019, 671: 1101-1107
[76] Mercier A, Gravouil K, Aucher W, et al. Fate of eight different polymers under uncontrolled composting conditions: Relationships between deterioration, biofilm formation, and the material surface properties [J]. Environmental Science & Technology, 2017, 51(4): 1988-1997
[77] Xie H F, Chen J J, Feng L M, et al. Chemotaxis-selective colonization of mangrove rhizosphere microbes on nine different microplastics [J]. Science of the Total Environment, 2021, 752: 142223
[78] Rosato A, Barone M, Negroni A, et al. Microbial colonization of different microplastic types and biotransformation of sorbed PCBs by a marine anaerobic bacterial community [J]. The Science of the Total Environment, 2020, 705: 135790
[79] Kirstein I V, Kirmizi S, Wichels A, et al. Dangerous hitchhikers? Evidence for potentially pathogenic Vibrio spp. on microplastic particles [J]. Marine Environmental Research, 2016, 120: 1-8
[80] Balasubramanian V, Natarajan K, Hemambika B, et al. High-density polyethylene (HDPE)-degrading potential bacteria from marine ecosystem of Gulf of Mannar, India [J]. Letters in Applied Microbiology, 2010, 51(2): 205-211
[81] Tribedi P, Gupta A D, Sil A K. Adaptation of Pseudomonas sp. AKS2 in biofilm on low-density polyethylene surface: An effective strategy for efficient survival and polymer degradation [J]. Bioresources and Bioprocessing, 2015, 2: 14
[82] Arkatkar A, Juwarkar A A, Bhaduri S, et al. Growth of Pseudomonas and Bacillus biofilms on pretreated polypropylene surface [J]. International Biodeterioration & Biodegradation, 2010, 64(6): 530-536
[83] Bhardwaj H, Gupta R, Tiwari A. Communities of microbial enzymes associated with biodegradation of plastics [J]. Journal of Polymers and the Environment, 2013, 21(2): 575-579
[84] Yang Y Y, Liu G H, Song W J, et al. Plastics in the marine environment are reservoirs for antibiotic and metal resistance genes [J]. Environment International, 2019, 123: 79-86
[85] Imran M, Das K R, Naik M M. Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: An emerging health threat [J]. Chemosphere, 2019, 215: 846-857
[86] Chen X C, Chen X F, Zhao Y H, et al. Effects of microplastic biofilms on nutrient cycling in simulated freshwater systems [J]. Science of the Total Environment, 2020, 719: 137276
[87] Arias-Andres M, Rojas-Jimenez K, Grossart H P. Collateral effects of microplastic pollution on aquatic microorganisms: An ecological perspective [J]. TrAC Trends in Analytical Chemistry, 2019, 112: 234-240