三唑酮是一种三唑类杀菌剂,占全球三唑类杀菌剂消费量[1]的30%以上。三唑酮在水环境中的普遍存在及其对水生生物的潜在不利影响已经引起广泛关注。由于农药的有效利用率极低[2],大量残留的三唑酮进入水环境,其使用区域的临近水体检测出了较高浓度的三唑酮[1,3]。在中国的九龙江、太湖和白洋淀等地均有三唑酮的检出,最高检出浓度为12 μg·L-1[3-6]。三唑酮具有稳定性强,流动性和吸附性好等特点,可在水生生物体内累积并发生生物转化[7-9],对水生生物产生毒性效应。例如,三唑酮可影响生物体的发育[10-12]、繁殖[13-14],引起神经毒性[15]、肝毒性[16]、心血管毒性[17]和氧化应激等[18-19]。三唑酮主要影响两栖动物的变态发育,农药的使用可能是两栖动物种群数量下降的原因之一[20]。三唑酮通过改变下丘脑-垂体-甲状腺轴(hypothalamic-pituitary-thyroid, HPT)相关基因的表达来干扰甲状腺激素的分泌,影响胚胎的孵化率,降低鱼类[13]和两栖动物[10]的繁殖能力。三唑酮影响两栖动物和鱼类生殖能力的另一途径是抑制睾酮向雌二醇的转化[12,21]。目前,关于三唑酮的毒性机制研究主要集中在高营养级的脊椎动物,如两栖动物和鱼类,而其对低营养级水生无脊椎动物的毒性效应研究相对较少,刘娜等[22]关于三唑酮毒性效应的研究发现三唑酮对大型溞蜕皮产生毒性作用,且其敏感性高于生长和繁殖,并进一步整理出三唑酮的有害结局路径[23],结果表明三唑酮可能通过抑制P450酶的活性从而降低甲壳类动物蜕皮激素的活性。目前仍缺少关于三唑酮对大型溞代际影响的研究以及分子机制上的探究。
由于基因组序列的可获得性、较短的生命周期和易于处理等优点,大型溞成为生态毒性基因组学研究的优秀模式生物[24],可用于评估化学品对水生生态系统的毒性影响[25]。大型溞处于藻-溞-鱼水环境食物链的中间位置,占据相对重要的生态位[26],因此,有关大型溞的毒性效应研究可能对整个水生环境食物链具有一定的参考作用。
本研究在整理归纳三唑酮的毒性效应和机制的基础上,开展大型溞的多代试验,探索三唑酮对大型溞的生长和繁殖毒性以及相应的毒性作用机制和途径。试验期间记录并分析大型溞的体长及产溞数量等指标,使用HiSeq平台进行转录组测序,分析处理组与对照组的差异表达基因及其功能富集通路,为探索三唑酮对水生无脊椎动物的毒性机制提供参考。
1 材料与方法(Materials and methods)
1.1 培养条件
试验所用大型溞为本实验室(中国环境监测总站)连续稳定培养5代以上的单克隆品系,个体之间差异较小。按照OECD 211 (Organization for Economic Co-Operation and Development)[27]的要求,大型溞培养使用Elendt M4培养基(配制所需化学药品均购自上海国药集团化学试剂有限公司),置于恒温培养箱中,培养温度为(21±1) ℃,每天光照16 h,黑暗8 h,光照强度在1 110~1 480 lx。每日以羊角月牙藻喂养,投饵密度为3.0×105~4×105 cells·mL-1。藻类按照OECD 201[28]要求培养,培养箱温度设为(23±1) ℃,每天光照10 h,黑暗8 h,光照强度在4 440~8 880 lx,培养4~5 d后在5 000 r·min-1转速下离心浓缩5 min,使用灭菌水稀释以降低营养盐浓度。
1.2 试验设计
三唑酮购于阿拉丁有限公司(Aladdin,中国上海),CAS号为43121-43-3,纯度为99.8%。丙酮购自康林科技有限公司(中国北京)。根据ECOTOX数据库(https:// cfpub.epa.gov/ecotox/search.cfm)中三唑酮毒性效应浓度和以往研究基础[22],设置6个浓度梯度,分别为5、12.5、25、50、100和200 μg·L-1(浓度从低到高依次标记为3~8),每个浓度设置5个重复。以丙酮(<0.1 mL·L-1)为助溶剂,使用M4培养基配制三唑酮母液。试验空白对照分别为M4培养基和含有0.02 mL·L-1丙酮的M4培养基(分别标记为1和2)。对照组未检出三唑酮。
采用图1中描述的试验设计进行三唑酮暴露的多代试验(F0、F1、F2)。试验开始前,选取20只个体大、游泳能力强且怀卵多的母溞置于1 L装有M4培养基的烧杯中,驯化培养。试验开始时,随机选取上述母溞所产<24 h的第三胎幼溞32只分别置于含有80 mL暴露液的100 mL烧杯中,每个烧杯一只溞,记为F0代。F0代所产<24 h的第三胎幼溞32只被选作子代F1,F1代所产<24 h的第三胎幼溞32只被选作子代F2。选择第三胎是因为早期的幼溞(第一胎和第二胎)存活不稳定。每代按照OECD 211[27]慢性毒性试验标准进行21 d半静态暴露试验,每天更换培养液取出新生儿并记录F0代大型溞的每胎产溞数量和时间以及每代相隔时间,暴露21 d后在显微镜下测定大型溞体长(不包括尾刺),并取1 mL暴露液过0.22 μm滤膜,随后保存在-20 ℃下待测。F1代和F2代出生7 d后进行转录组测试和基因表达分析。
1.3 三唑酮暴露液浓度检测
暴露21 d后取暴露液1 mL,经过前处理去除杂质和影响物质,使用LC-MS/MS-8040 (岛津,日本)检测暴露液中三唑酮实际浓度。液相色谱柱为ACQUITY UPLC® BEH C18 (1.7 μm, 50 mm×2.1 mm, Waters, MA, USA),进样量为5 μL,流动相由0.1%甲酸水溶液和乙腈组成。流动相流速为0.3 mL·min-1,柱温为40 ℃。洗脱梯度如表1所示。
定量分析使用LC-MS/MS-8040 (岛津,日本)完成,采用多反应监测(multiple reaction monitoring, MRM)与电喷雾正离子源(ESI+)模式。氮气作为脱溶剂和雾化气。母离子294 (m/z),子离子197 (m/z)。名义浓度为5.00、12.50、25.00、50.00、100.00和200.00 μg·L-1的三唑酮暴露液实测浓度分别为4.72、11.36、32.02、63.24、116.79和229.00 μg·L-1。
1.4 转录组测序
每个处理组随机挑选约40只大型溞作为一个生物学重复,使用TRIzol(Invitrogen,美国)提取大型溞总RNA,并使用DNaseⅠ(TaKara,大连,中国)去除基因组DNA。利用2100 Bioanalyser(Agilent,美国)、ND-2000微量分光光度计(Thermo Scientific,美国)对所提取RNA的浓度和纯度进行检测,保证所有RNA样品的完整性(OD260/OD280=1.8~2.2, OD260/OD230≥2.0, RIN≥8.5, 28S/18S≥1.0)并进行转录组测序。mRNA测序基于HiSeq平台,采用Illumina TruSeqTM RNA sample prep Kit方法进行文库构建,主要操作流程为:提取总RNA(>1 μg),富集mRNA,mRNA片段化,反转成cDNA,adaptor连接,Illumina测序。
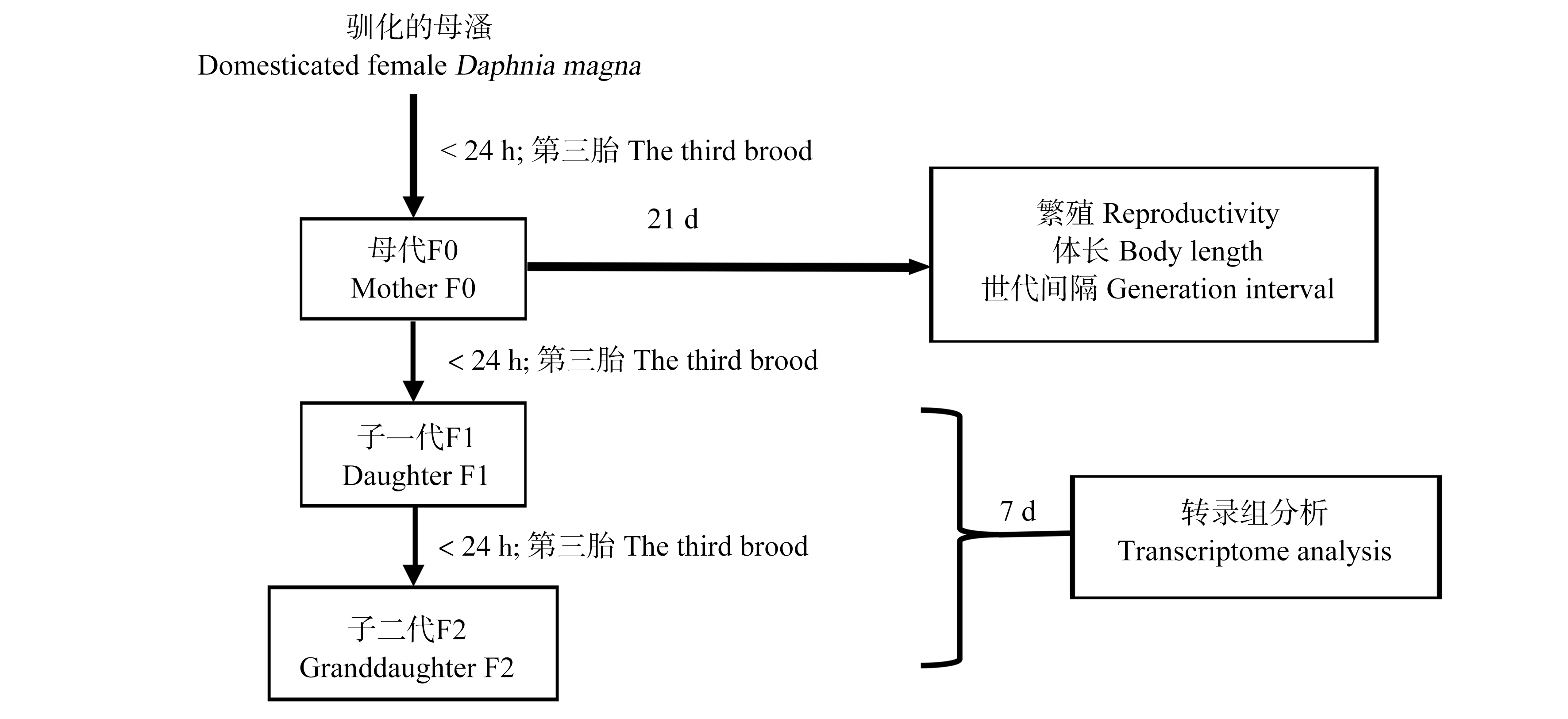
图1 多代试验示意图
Fig. 1 Diagram of multi-generational experiment
1.5 差异表达基因
使用RSEM(https://deweylab.biostat.wisc.edu/resm/)对基因的表达水平进行定量分析,获得基因/转录本的Read Counts数后,使用DEGseq (http://bioconductor.org/packages/stats/bioc/DESeq/)软件进行样本间基因的表达差异分析,以FPKM(fragments per kilobases per millionreads)衡量差异表达的显著性,使用错误发现率(false discovery rate, FDR)和差异倍数(fold change, FC)作为评判标准,当一个基因同时满足FDR<0.05和|log2FC|>1时,视该基因为差异表达基因(differentially expressed gene, DEG)。
1.6 差异表达基因的KEGG富集
使用KOBAS(https:// kobas.cbi.pku.edu.cn/home.do)对不同组别的差异表达基因进行功能富集分析,在基因功能水平上阐述三唑酮暴露对大型溞可能的作用机制和途径。利用KEGG数据库,可将基因按照参与的通路或行使的功能分类,能够识别出与生物现象最相关的生物学过程。使用BH(Benjamini and Hochberg)方法进行多重检验矫正P值,P≤0.05,认为此GO富集功能或KEGG通路存在显著富集情况。
1.7 统计分析
本研究涉及的数据统计分析使用Origin 2021(OriginLab Corporation, Northampton, MA, USA),The R Programming Language 4.0.3和IBM SPSS Statistics 26(BM Corporation, Armonk, NY, USA)。使用单因素方差分析进行显著性检验和方差同质性检验,重复组取算术平均值,表示为平均值±标准差(standard deviation, SD)。P<0.05表示两样本间存在显著性差异。
2 结果(Results)
2.1 三唑酮对大型溞F0代生长和繁殖的影响
F0代的体长是大型溞的生长指标,暴露结束后,在显微镜下测定不同三唑酮暴露浓度下大型溞的体长(不包括尾刺),结果显示,仅在200 μg·L-1浓度时体长明显降低(P=0.000),其他浓度下体长变化不显著(图2(a))。本研究大型溞为孤雌生殖,将F0代大型溞21 d平均每胎产溞量作为大型溞的繁殖指标,结果显示,在浓度为100 μg·L-1和200 μg·L-1时21 d平均每胎产溞量均明显降低(P=0.014, P=0.000),而其他浓度下的繁殖能力差异不明显(图2(b))。除体长和繁殖外,本研究也记录了不同浓度下F0代产下一代F1以及F1代产下一代F2所需要的时间间隔。通过组间方差分析发现,浓度为5、12.5、25、50和100 μg·L-1时F0从出生到产下F1的间隔时间和F1从出生到产下F2的间隔时间增加不明显,而当浓度为200 μg·L-1时两代世代间隔时间均显著增加(P=0.008,P=0.000)(表2)。比较不同浓度下两代世代间隔时间的差异可知(表2),与对照组相比,12.5 μg·L-1(P=0.033)和100 μg·L-1(P=0.008)浓度处理下世代间隔时间显著增加。
2.2 差异表达基因
F1和F2代大型溞在三唑酮中暴露7 d后,进行转录组测试分析,共获得162.94 Gb Clean Data,且各样品Clean Data均达到6.38 Gb以上,Q30碱基百分比在93.5%以上。转录组测试共检测到20 187个基因。分别将各样品的Clean Reads与指定的参考基因组(https://www.ncbi.nlm.nih.gov/gen-ome/10953?genome_assembly_id=434305)进行序列比对,比对率为79.02%~91.76%。基于各浓度下基因表达量,进行样本间相关性分析和PCA分析,分别如图3和图4所示,样本间相关性分析表明样本间相关性较高,PCA分析可以发现高浓度(200 μg·L-1)处理组与其他浓度组间有较大差异,同时,F1代和F2代间的转录组响应也存在显著差异。因此我们对F1代、F2代以及F1 vs F2的基因表达情况进行了进一步分析。
表1 目标物质的洗脱梯度
Table 1 Elution gradient of target substance

时间/minTime/min流速/(mL·min-1)Flow rate/(mL·min-1)流动相/%Mobile phase/%0.1%甲酸水溶液0.1% Formic acid乙腈Acetonitrile进样量/μLInjection volume/μL00.395550~0.50.395550.5~0.60.3257550.6~2.00.329852.0~3.00.329853.0~3.10.395553.1~4.00.39555
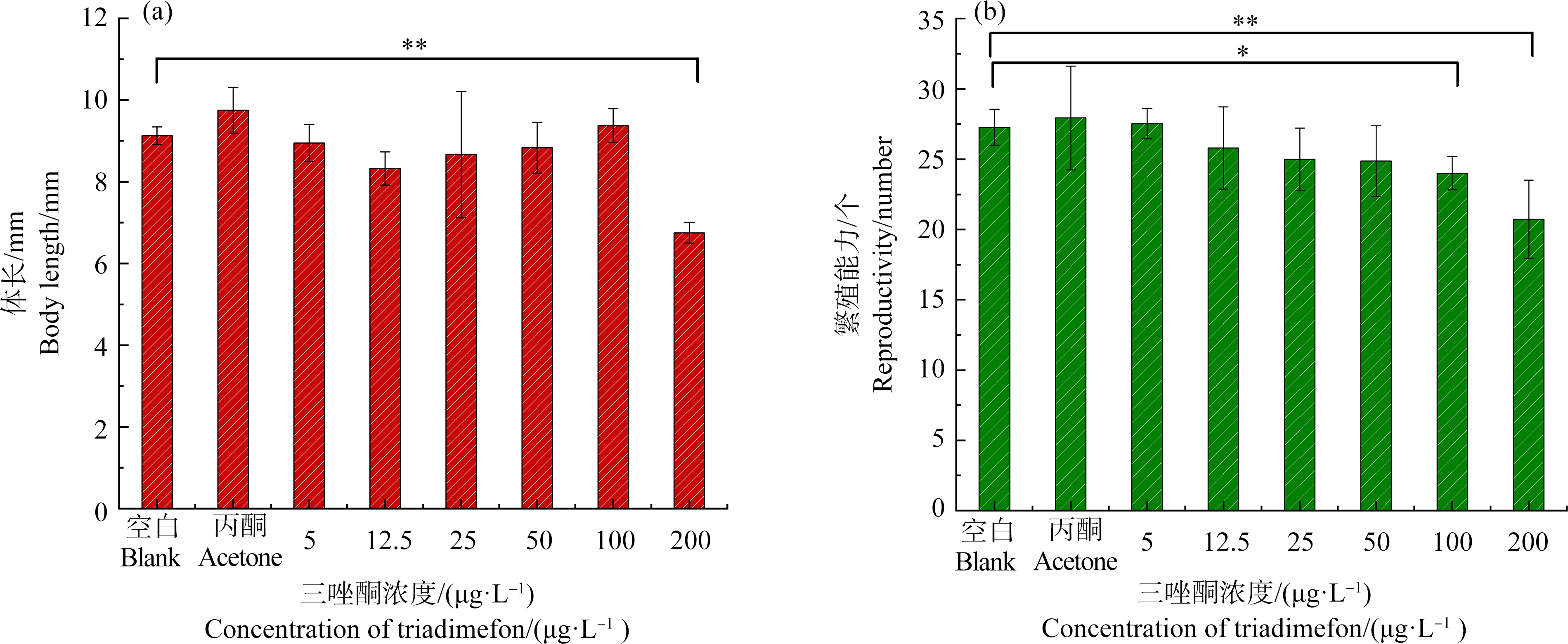
图2 三唑酮暴露21 d对大型溞F0代体长(a)和繁殖(b)的影响
注:数据表示为平均值±标准差,*代表P<0.05,**代表P<0.01,与对照组相比有显著性差异。
Fig. 2 Influences of triadimefon 21 d exposure on length (a) and reproduction (b) of Daphnia magna (F0)
Note: Data was represented by mean±standard deviation; compared with control, the exposure group had significant difference at
*P<0.05, **P<0.01 level.
表2 三唑酮暴露对大型溞世代间隔时间的影响
Table 2 Influences of triadimefon exposure on generation interval time of Daphnia magna
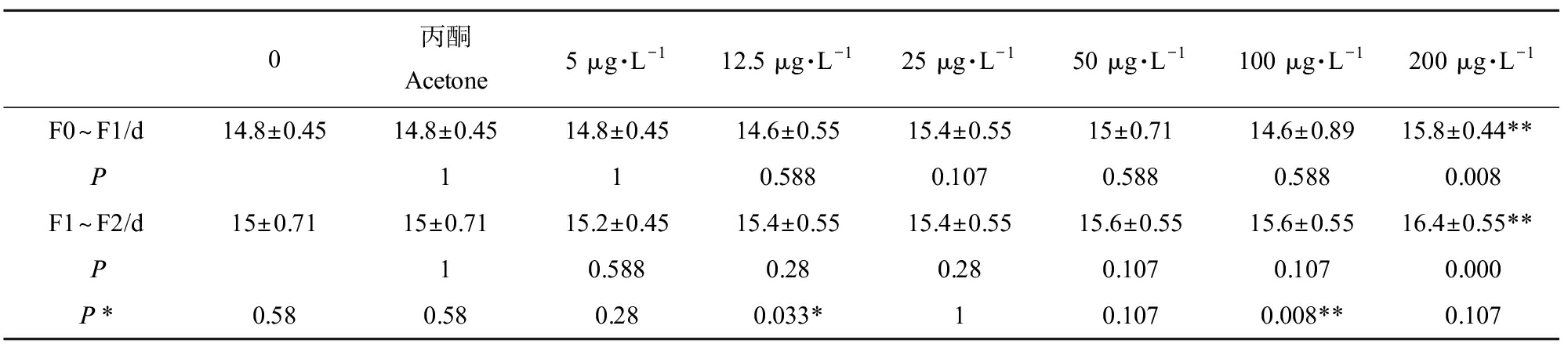
0丙酮Acetone5 μg·L-112.5 μg·L-125 μg·L-150 μg·L-1100 μg·L-1200 μg·L-1F0~F1/d14.8±0.4514.8±0.4514.8±0.4514.6±0.5515.4±0.5515±0.7114.6±0.8915.8±0.44**P110.5880.1070.5880.5880.008F1~F2/d15±0.7115±0.7115.2±0.4515.4±0.5515.4±0.5515.6±0.5515.6±0.5516.4±0.55**P10.5880.280.280.1070.1070.000P *0.580.580.280.033*10.1070.008**0.107
注:世代间隔时间为从F0出生到F0产下第三胎(即F1),以此类推;数据表示为平均值±标准差,*代表P<0.05,**代表P<0.01,与对照组相比有显著性差异,P *表示不同浓度下两代世代间隔时间两两比对的差异,*代表P *<0.05,**代表P *<0.01。
Note: One generation is from F0 birth to F0 giving birth to the third brood (F1), and so forth; data was represented by mean±standard deviation; compared with control, the exposure group had significant difference at *P<0.05, **P<0.01 level; P *represents differences between generation interval time of F0~F1 and F1~F2 under different exposure at *P *<0.05, **P *<0.01 level.
基于上述表达量分析,F1代对照组(空白对照和溶剂对照)与处理组(试验设置的6个浓度组)的差异表达基因共376个,其中上调基因322个,下调基因54个(图4(a))。F2代对照组与处理组的差异表达基因共422个,其中上调基因87个,下调基因335个(图4(b))。而F1和F2代的差异表达基因共2 604个,其中上调基因1 101个,下调基因1 503个(图4(c)),由火山图可知,F1代上调基因明显高于下调基因,而F2代则相反。相比之下,F1代与F2代比较的差异表达基因上调与下调基因数基本相似。F1代对照组(空白对照和溶剂对照)与所有处理组(试验设置的6个浓度组)共有差异表达基因36个,F2代对照组与所有处理组共有差异表达基因82个,将F1代的基因与F2代的基因对比得到两者共有差异表达基因共336个(如图4(d)~(f)所示)。
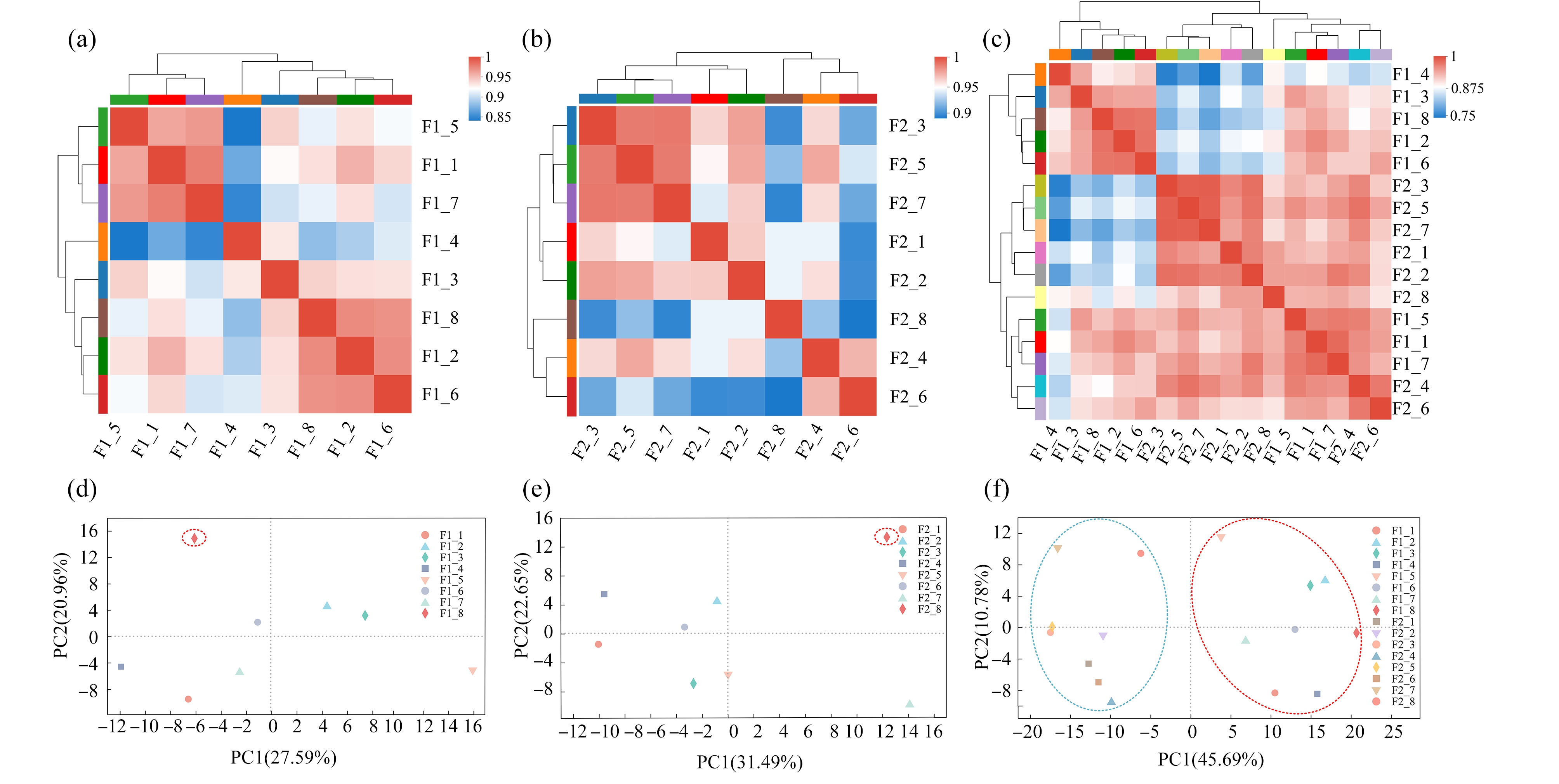
图3 样本间相关性分析和PCA分析
注:F1_1~F1_8分别代表F1代空白对照组、丙酮对照组以及5、12.5、25、50、100、200 μg·L-1三唑酮处理组;F2_1~F2_8分别代表F2代
空白对照组、丙酮对照组以及5、12.5、25、50、100、200 μg·L-1三唑酮处理组;(d)和(e)中红色虚线圈出是200 μg·L-1处理组,
(f)中红色虚线圈出是F1代,蓝色虚线圈出是F2代。
Fig. 3 Correlation analysis and PCA analysis among samples
Note: F1_1~F1_8 represent control group, acetone control group, and 5, 12.5, 25, 50, 100, 200 μg·L-1 triadimefon treatment group, respectively;
F2_1~F2_8 represent control group, acetone control group, and 5, 12.5, 25, 50, 100, 200 μg·L-1 μg·L-1 triadimefon treatment group; circled by a red
dotted line in (d) and (e) is treatment group of 200 μg·L-1; circled by a red dotted line and a blue dotted line in (f) represent F1 and F2, respectively.
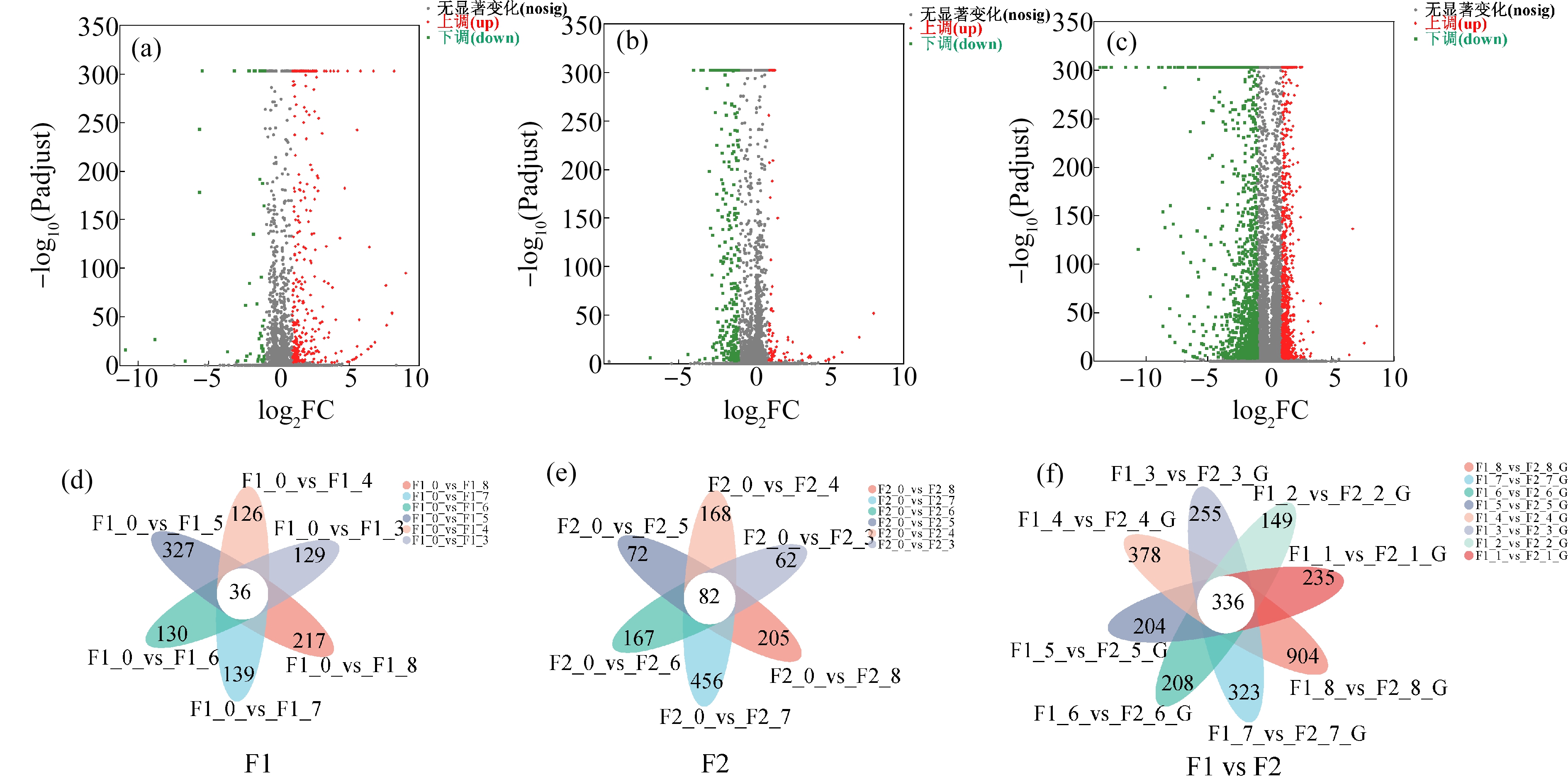
图4 差异表达基因火山图(a)、(b)、(c)和Venn图(d)、(e)、(f)
注:F1_0和F2_0分别代表F1代和F2代对照组,F1_3~F1_8分别代表F1代5、12.5、25、50、100、200 μg·L-1三唑酮处理组;
F2_3~F2_8分别代表F2代5、12.5、25、50、100、200 μg·L-1三唑酮处理组。
Fig. 4 Volcano figures of differentially expressed genes (a), (b), (c) and Venn figures of differentially expressed genes (d), (e), (f)
Note: F1_0 and F2_0 represent control group in F1 and F2, respectively; F1_3~F1_8 represent 5, 12.5, 25, 50, 100, 200 μg·L-1 triadimefon
treatment group in F1, respectively; F2_3~F2_8 represent 5, 12.5, 25, 50, 100, 200 μg·L-1 triadimefon treatment group in F2.
2.3 关键通路和关键基因
通过上述基因表达差异的分析,对F1代和F2代的所有差异表达基因进行功能聚类和通路分析(表3和表4)。结果表明,F1代差异表达基因显著富集的主要通路包括甾类激素生物合成、视黄醇代谢、酮体的合成与降解等代谢通路、氧化应激、内质网应激以及与寿命调节和消化系统相关的通路。其中寿命调节和蛋白质消化吸收是富集最为显著的通路。F2代显著富集通路主要有酮体的合成与降解、类固醇生物合成和谷胱甘肽代谢等代谢通路、抗原的处理和呈递以及造血细胞谱系等免疫系统通路、内分泌系统和氮代谢。其中氮代谢和造血细胞谱系是富集最显著的通路。对富集到关键通路的显著差异表达基因在不同浓度水平下进行表达量分析(图5)。F1代中富集到关键通路的绝大部分关键基因在浓度100 μg·L-1和200 μg·L-1时显著上调(图5),仅基因trypsin alpha-1-like和基因zinc metalloproteinase nas-12-like在浓度200 μg·L-1时显著下调,且其他与蛋白质消化吸收相关的基因在该浓度下与对照组之间无显著差异,而在较低浓度时上调。这说明在较低浓度下,机体通过上调蛋白质消化吸收相关基因应对外界胁迫,而200 μg·L-1可能已经超过其基因水平的毒性阈值,从而引起不利的基因表达。而与药物代谢相关的CYP3A (CYP3A是一种重要的CYP450酶系,可催化许多外源药物的代谢)和UGT(葡萄糖醛酸转移酶)相关基因的表达在不同浓度下呈现不同的表达模式,这些基因富集在甾类激素生物合成通路,这可能导致内分泌干扰作用[29-30]。3-羟基丁酸脱氢酶、全反式视黄醇13,14还原酶相关基因在各处理浓度下均上调。不同于F1代,在F2代中,大部分关键基因显著下调。LIPA(脂肪酶)、CD13(氨肽酶类)、CD63(四磷酸腺苷)相关基因在高浓度处理组(50、100和200 μg·L-1)显著下调,CA(碳酸酐酶)、CTSL(组织蛋白酶L)和BDH(3-羟基丁酸脱氢酶)相关基因在各处理组显著下调。F1代和F2代基因表达的差异说明F1代的暴露会影响其子代的基因表达,加剧对子代的毒性作用。
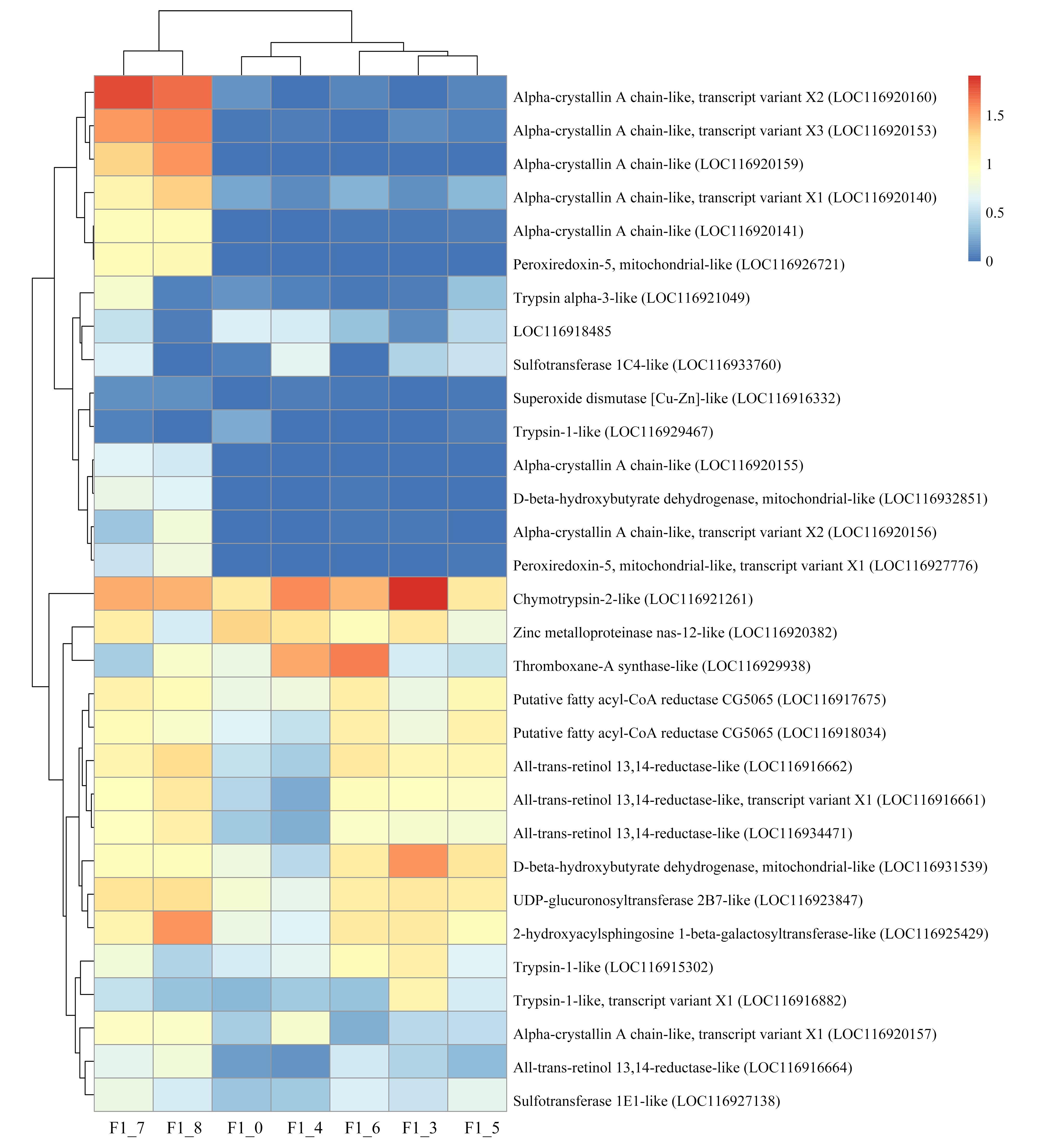
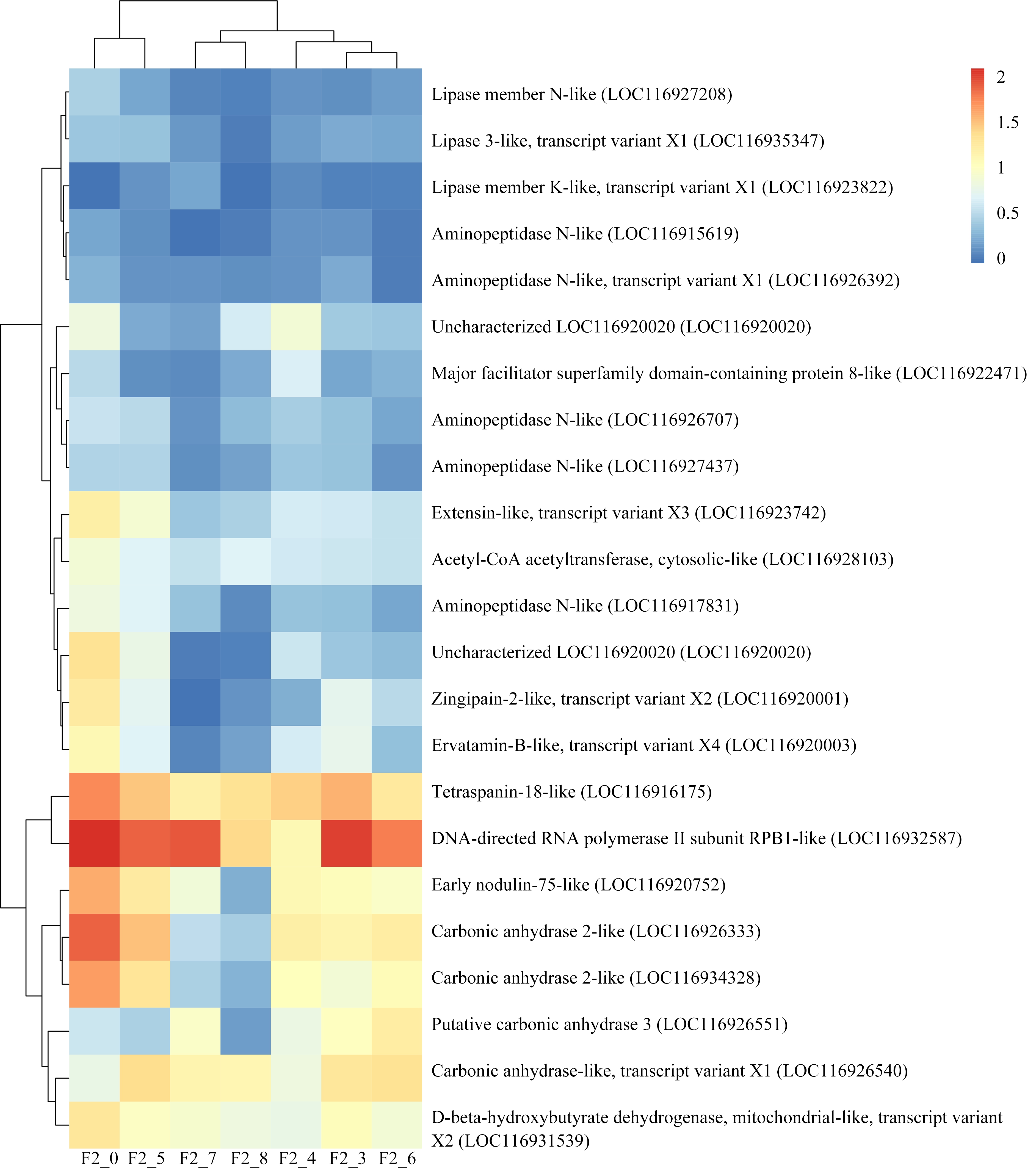
图5 关键差异表达基因表达量(log(TPM+1))分布图
注:F1_0和F2_0分别代表F1代和F2代对照组,F1_3~F1_8分别代表F1代5、12.5、25、50、100、200 μg·L-1三唑酮处理组;
F2_3~F2_8分别代表F2代5、12.5、25、50、100、200 μg·L-1三唑酮处理组。
Fig. 5 Expression distribution (log(TPM+1)) of key differentially expressed genes
Note: F1_0 and F2_0 represent control group in F1 and F2, respectively; F1_3~F1_8 represent 5, 12.5, 25, 50, 100, 200 μg·L-1 triadimefon
treatment group in F1, respectively; F2_3~F2_8 represent 5, 12.5, 25, 50, 100, 200 μg·L-1 triadimefon treatment group in F2.
表3 F1代差异表达基因KEGG富集主要信号通路
Table 3 Main pathways of KEGG enrichment of differentially expressed genes in F1
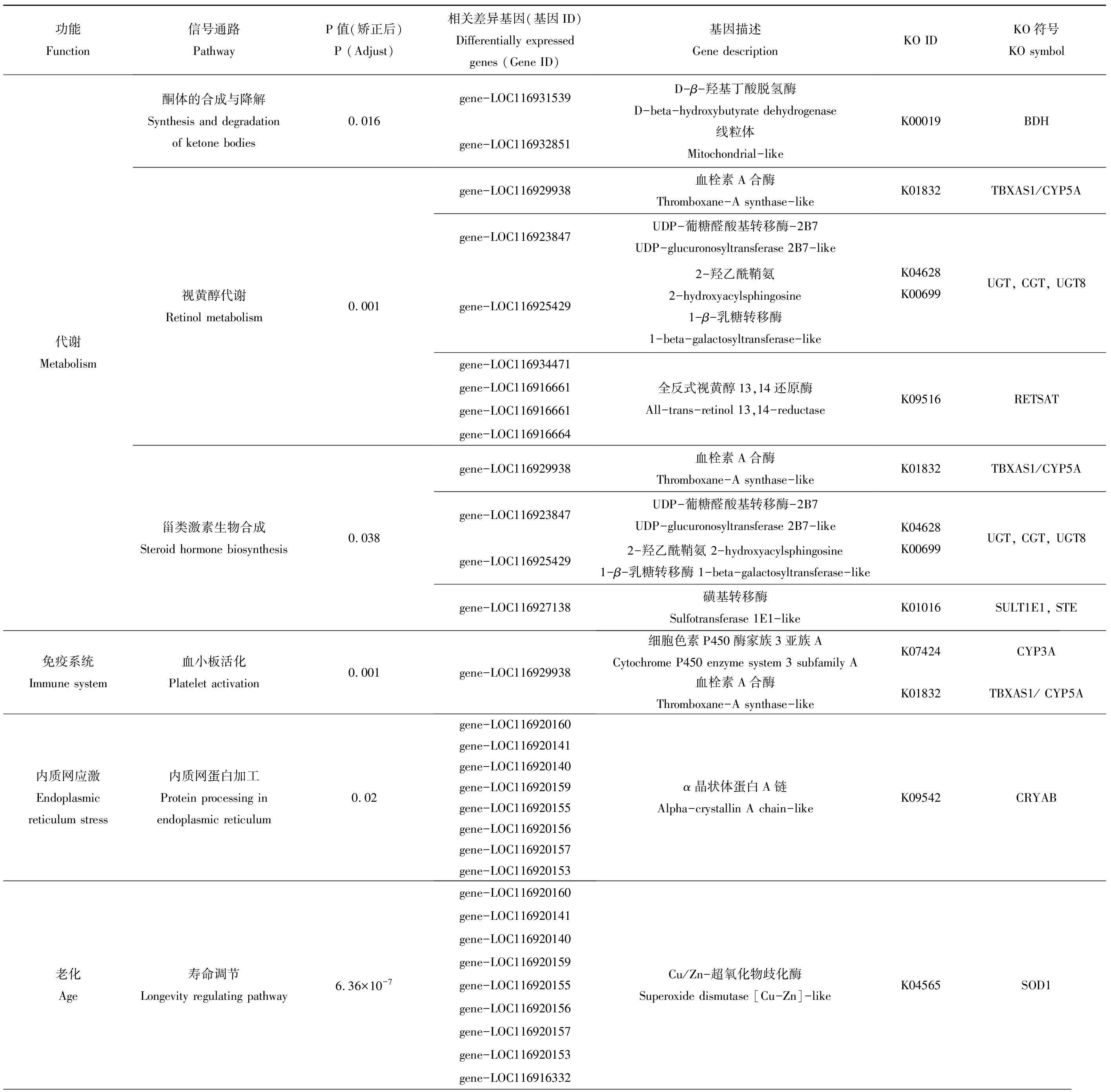
功能Function信号通路PathwayP值(矫正后)P (Adjust)相关差异基因(基因ID)Differentially expressed genes (Gene ID)基因描述Gene descriptionKO IDKO符号KO symbol代谢Metabolism酮体的合成与降解Synthesis and degradation of ketone bodies0.016视黄醇代谢Retinol metabolism0.001甾类激素生物合成Steroid hormone biosynthesis0.038gene-LOC116931539gene-LOC116932851gene-LOC116929938gene-LOC116923847gene-LOC116925429gene-LOC116934471gene-LOC116916661gene-LOC116916661gene-LOC116916664gene-LOC116929938gene-LOC116923847gene-LOC116925429gene-LOC116927138D-β-羟基丁酸脱氢酶D-beta-hydroxybutyrate dehydrogenase线粒体Mitochondrial-like血栓素A合酶Thromboxane-A synthase-likeUDP-葡糖醛酸基转移酶-2B7UDP-glucuronosyltransferase 2B7-like2-羟乙酰鞘氨2-hydroxyacylsphingosine1-β-乳糖转移酶1-beta-galactosyltransferase-like全反式视黄醇13,14还原酶All-trans-retinol 13,14-reductase血栓素A合酶Thromboxane-A synthase-likeUDP-葡糖醛酸基转移酶-2B7UDP-glucuronosyltransferase 2B7-like2-羟乙酰鞘氨 2-hydroxyacylsphingosine1-β-乳糖转移酶 1-beta-galactosyltransferase-like磺基转移酶Sulfotransferase 1E1-likeK00019BDHK01832TBXAS1/CYP5AK04628K00699UGT, CGT, UGT8K09516RETSATK01832TBXAS1/CYP5AK04628K00699UGT, CGT, UGT8K01016SULT1E1, STE免疫系统Immune system血小板活化Platelet activation0.001gene-LOC116929938细胞色素P450酶家族3亚族ACytochrome P450 enzyme system 3 subfamily AK07424CYP3A血栓素A合酶Thromboxane-A synthase-likeK01832TBXAS1/ CYP5A内质网应激Endoplasmic reticulum stress内质网蛋白加工Protein processing in endoplasmic reticulum0.02gene-LOC116920160gene-LOC116920141gene-LOC116920140gene-LOC116920159gene-LOC116920155gene-LOC116920156gene-LOC116920157gene-LOC116920153α晶状体蛋白A链Alpha-crystallin A chain-likeK09542CRYAB老化Age寿命调节Longevity regulating pathway6.36×10-7gene-LOC116920160gene-LOC116920141gene-LOC116920140gene-LOC116920159gene-LOC116920155gene-LOC116920156gene-LOC116920157gene-LOC116920153gene-LOC116916332Cu/Zn-超氧化物歧化酶Superoxide dismutase [Cu-Zn]-likeK04565SOD1
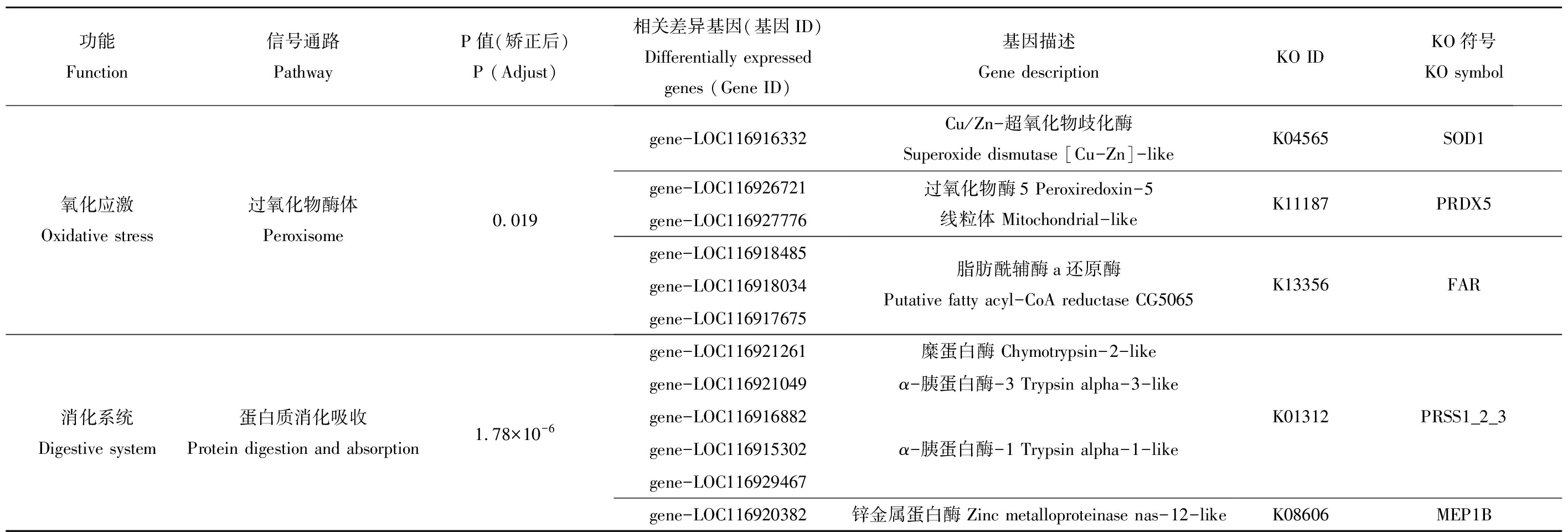
功能Function信号通路PathwayP值(矫正后)P (Adjust)相关差异基因(基因ID)Differentially expressed genes (Gene ID)基因描述Gene descriptionKO IDKO符号KO symbol氧化应激Oxidative stress过氧化物酶体Peroxisome0.019gene-LOC116916332gene-LOC116926721gene-LOC116927776gene-LOC116918485gene-LOC116918034gene-LOC116917675Cu/Zn-超氧化物歧化酶Superoxide dismutase [Cu-Zn]-likeK04565SOD1过氧化物酶5 Peroxiredoxin-5线粒体 Mitochondrial-likeK11187PRDX5脂肪酰辅酶a还原酶Putative fatty acyl-CoA reductase CG5065K13356FAR消化系统Digestive system蛋白质消化吸收Protein digestion and absorption1.78×10-6gene-LOC116921261gene-LOC116921049gene-LOC116916882gene-LOC116915302gene-LOC116929467gene-LOC116920382糜蛋白酶 Chymotrypsin-2-likeα-胰蛋白酶-3 Trypsin alpha-3-likeα-胰蛋白酶-1 Trypsin alpha-1-like锌金属蛋白酶 Zinc metalloproteinase nas-12-likeK01312PRSS1_2_3K08606MEP1B
表4 F2代差异基因KEGG富集主要信号通路
Table 4 Main pathways of KEGG enrichment of differentially expressed genes in F2
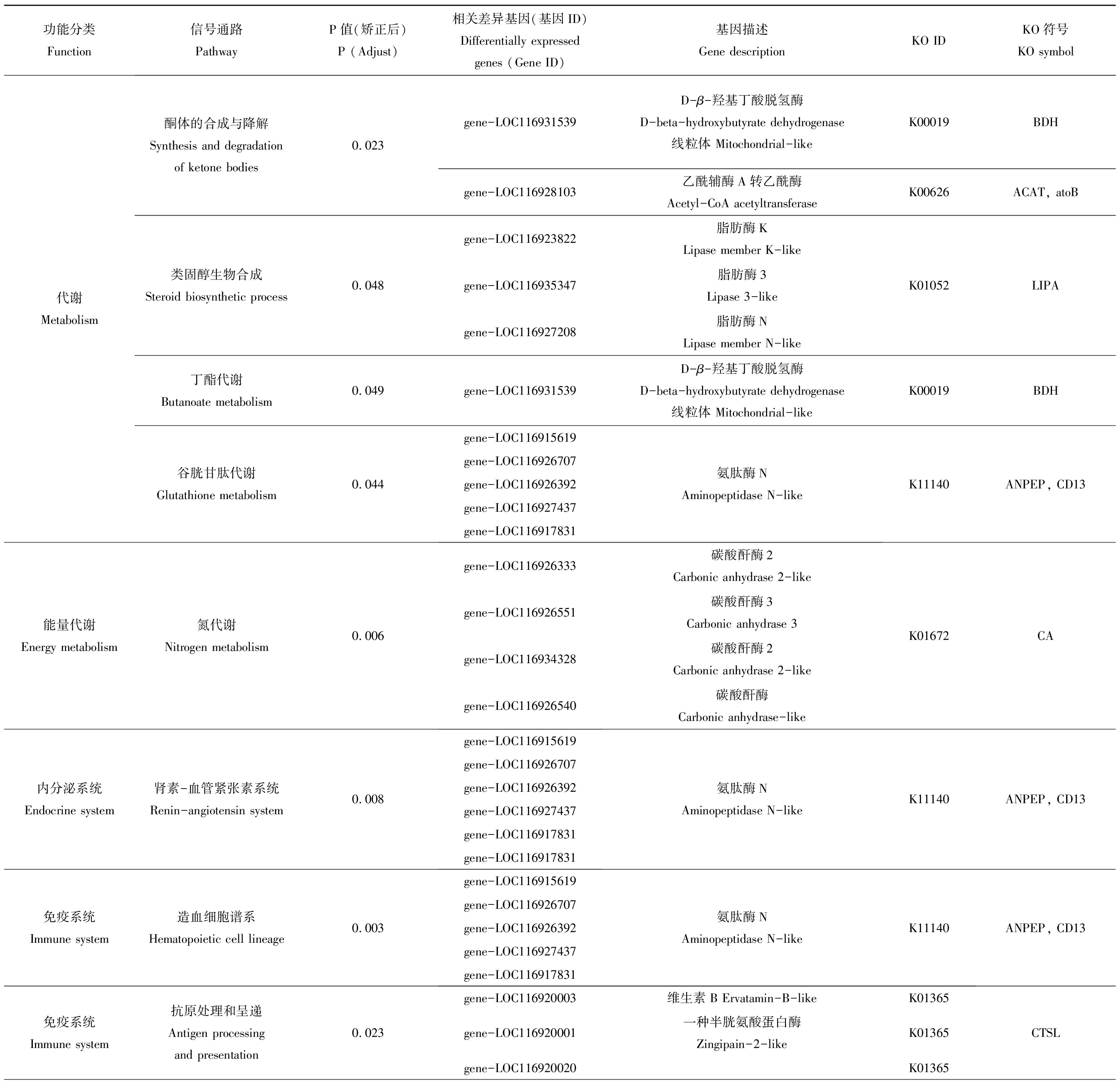
功能分类Function信号通路PathwayP值(矫正后)P (Adjust)相关差异基因(基因ID)Differentially expressed genes (Gene ID)基因描述Gene descriptionKO IDKO符号KO symbol代谢Metabolism酮体的合成与降解Synthesis and degradation of ketone bodies0.023类固醇生物合成Steroid biosynthetic process0.048丁酯代谢Butanoate metabolism0.049谷胱甘肽代谢Glutathione metabolism0.044gene-LOC116931539gene-LOC116928103gene-LOC116923822gene-LOC116935347gene-LOC116927208gene-LOC116931539gene-LOC116915619gene-LOC116926707gene-LOC116926392gene-LOC116927437gene-LOC116917831D-β-羟基丁酸脱氢酶D-beta-hydroxybutyrate dehydrogenase线粒体 Mitochondrial-like乙酰辅酶A转乙酰酶Acetyl-CoA acetyltransferase脂肪酶KLipase member K-like脂肪酶3 Lipase 3-like脂肪酶NLipase member N-likeD-β-羟基丁酸脱氢酶D-beta-hydroxybutyrate dehydrogenase线粒体 Mitochondrial-like氨肽酶NAminopeptidase N-likeK00019BDHK00626ACAT, atoBK01052LIPAK00019BDHK11140ANPEP, CD13能量代谢Energy metabolism氮代谢Nitrogen metabolism0.006gene-LOC116926333碳酸酐酶2Carbonic anhydrase 2-likegene-LOC116926551碳酸酐酶3Carbonic anhydrase 3gene-LOC116934328碳酸酐酶2Carbonic anhydrase 2-likegene-LOC116926540碳酸酐酶Carbonic anhydrase-likeK01672CA内分泌系统Endocrine system肾素-血管紧张素系统Renin-angiotensin system0.008gene-LOC116915619gene-LOC116926707gene-LOC116926392gene-LOC116927437gene-LOC116917831gene-LOC116917831氨肽酶NAminopeptidase N-likeK11140ANPEP, CD13免疫系统Immune system造血细胞谱系Hematopoietic cell lineage0.003gene-LOC116915619gene-LOC116926707gene-LOC116926392gene-LOC116927437gene-LOC116917831氨肽酶NAminopeptidase N-likeK11140ANPEP, CD13免疫系统Immune system抗原处理和呈递Antigen processing and presentation0.023gene-LOC116920003维生素B Ervatamin-B-likeK01365gene-LOC116920001一种半胱氨酸蛋白酶Zingipain-2-likeK01365gene-LOC116920020K01365CTSL
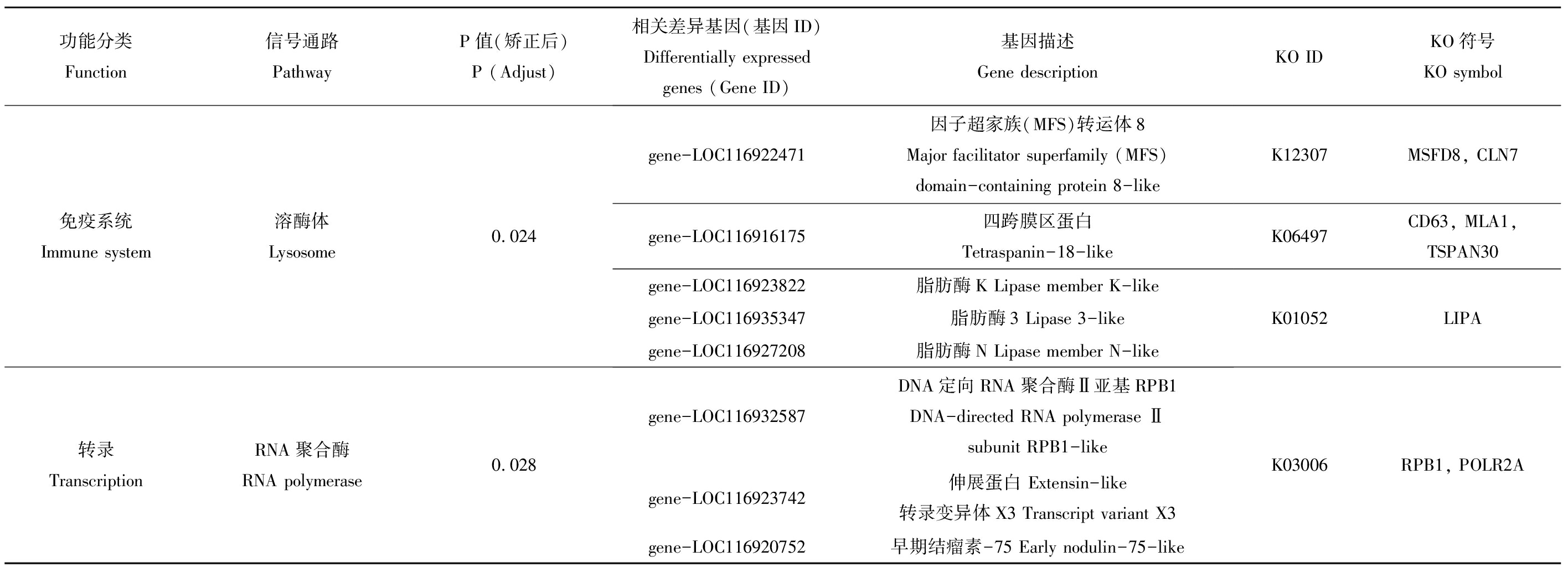
功能分类Function信号通路PathwayP值(矫正后)P (Adjust)相关差异基因(基因ID)Differentially expressed genes (Gene ID)基因描述Gene descriptionKO IDKO符号KO symbol免疫系统Immune system溶酶体Lysosome0.024gene-LOC116922471因子超家族(MFS)转运体8Major facilitator superfamily (MFS) domain-containing protein 8-likegene-LOC116916175四跨膜区蛋白Tetraspanin-18-likegene-LOC116923822脂肪酶K Lipase member K-likegene-LOC116935347脂肪酶3 Lipase 3-likegene-LOC116927208脂肪酶N Lipase member N-likeK12307MSFD8, CLN7K06497CD63, MLA1, TSPAN30K01052LIPA转录TranscriptionRNA聚合酶RNA polymerase0.028gene-LOC116932587DNA定向RNA聚合酶Ⅱ亚基RPB1DNA-directed RNA polymerase Ⅱ subunit RPB1-likegene-LOC116923742伸展蛋白 Extensin-like转录变异体X3 Transcript variant X3gene-LOC116920752早期结瘤素-75 Early nodulin-75-likeK03006RPB1, POLR2A
3 讨论(Discussion)
本研究依据OECD 211[27]测试方法开展大型溞多代实验,评估三唑酮对大型溞的毒性效应。结果表明:(1)三唑酮浓度为200 μg·L-1时,显著影响大型溞体长。刘娜等[22]探究了三唑酮对大型溞不同测试终点的毒性效应,发现当三唑酮浓度≤100 μg·L-1时,大型溞体长与对照组无明显差别,计算以体长为生长指标的最大无影响浓度(no observed effect concentration, NOEC)为100 μg·L-1;(2)当三唑酮浓度≥100 μg·L-1时,大型溞繁殖能力显著降低,说明该浓度可能已经达到造成繁殖毒性的阈值;(3)三唑酮的暴露可能导致世代间隔时间延长。胡方华等[31]关于三唑酮对大型溞的慢性毒性效应的研究表明,三唑酮对第2代大型溞染毒的影响比对第1代的影响更大。这说明三唑酮对大型溞存在长期毒性效应,有必要考虑三唑酮对大型溞的传代效应。
为进一步研究三唑酮对大型溞毒性效应的分子机制,本研究通过转录组分析对差异表达基因进行KEGG pathway分析,探索三唑酮对大型溞可能的毒性作用机制和途径。超氧化物歧化酶(SOD)是反映机体抗氧化反应水平的重要抗氧化酶,能催化![]() 分解为H2O2。当暴露于一定的环境压力时,机体会上调氧化应激酶的表达,以避免氧化损伤。然而,当氧化应激超过阈值时,抗氧化系统会被破坏,导致酶活性降低[32]。F1中SOD相关基因上调说明三唑酮暴露会引起大型溞的氧化应激反应且未达到阈值。在鱼类和两栖动物中也检测到膜脂的氧化应激和过氧化[18-19]。在人体中,CRYAB(α-B-晶状体蛋白)在许多神经系统疾病中过度表达[33],其相关基因在大型溞F1代中的表达量上调可能说明暴露于三唑酮的大型溞触发了抗凋亡和神经保护功能。因此三唑酮对大型溞可能具有神经毒性并引起细胞凋亡和损伤。以往的研究发现三唑酮能够与多巴胺转运体结合,抑制单胺的摄取,从而引起哺乳动物的神经毒性[34-35]。F1代中与CYP3A和CYP3A相关的基因表达在不同浓度下呈现不同模式(上调或下调),F2代中与BDH(酮体代谢过程中的关键基因)和LIPA相关基因表达下调,这些基因富集在视黄醇代谢、甾类激素生物合成、酮体的合成与降解、丁酯代谢和类固醇合成通路,且三唑酮在高浓度处理组影响F0代繁殖和体长,说明三唑酮可能产生内分泌干扰作用,先前的研究也发现三唑酮会通过影响HPT轴对鱼类和两栖动物产生内分泌干扰作用[10,21,36]。而三唑酮对大型溞的内分泌干扰机制尚不确定,可能是通过抑制CYP酶的活性来减少蜕皮激素的合成实现的[37]。除此之外,F2代主要信号通路是免疫反应,与之相关的基因(CD13、CD63和CTSL)显著下调。CD63是在多种真核生物中发现的跨膜蛋白,参与多种细胞过程,包括免疫反应、信号转导、运输等[38-39]。Jung等[40]研究了CD63在斑马鱼中的表达和功能,发现在斑马鱼胚胎的发育过程中,CD63在免疫系统器官如轴静脉和前肾管中表达。综上所述,短期暴露于三唑酮,可能引起大型溞的氧化应激和神经毒性,而长期暴露则可能会引起免疫毒性,内分泌干扰作用将一直存在。
分解为H2O2。当暴露于一定的环境压力时,机体会上调氧化应激酶的表达,以避免氧化损伤。然而,当氧化应激超过阈值时,抗氧化系统会被破坏,导致酶活性降低[32]。F1中SOD相关基因上调说明三唑酮暴露会引起大型溞的氧化应激反应且未达到阈值。在鱼类和两栖动物中也检测到膜脂的氧化应激和过氧化[18-19]。在人体中,CRYAB(α-B-晶状体蛋白)在许多神经系统疾病中过度表达[33],其相关基因在大型溞F1代中的表达量上调可能说明暴露于三唑酮的大型溞触发了抗凋亡和神经保护功能。因此三唑酮对大型溞可能具有神经毒性并引起细胞凋亡和损伤。以往的研究发现三唑酮能够与多巴胺转运体结合,抑制单胺的摄取,从而引起哺乳动物的神经毒性[34-35]。F1代中与CYP3A和CYP3A相关的基因表达在不同浓度下呈现不同模式(上调或下调),F2代中与BDH(酮体代谢过程中的关键基因)和LIPA相关基因表达下调,这些基因富集在视黄醇代谢、甾类激素生物合成、酮体的合成与降解、丁酯代谢和类固醇合成通路,且三唑酮在高浓度处理组影响F0代繁殖和体长,说明三唑酮可能产生内分泌干扰作用,先前的研究也发现三唑酮会通过影响HPT轴对鱼类和两栖动物产生内分泌干扰作用[10,21,36]。而三唑酮对大型溞的内分泌干扰机制尚不确定,可能是通过抑制CYP酶的活性来减少蜕皮激素的合成实现的[37]。除此之外,F2代主要信号通路是免疫反应,与之相关的基因(CD13、CD63和CTSL)显著下调。CD63是在多种真核生物中发现的跨膜蛋白,参与多种细胞过程,包括免疫反应、信号转导、运输等[38-39]。Jung等[40]研究了CD63在斑马鱼中的表达和功能,发现在斑马鱼胚胎的发育过程中,CD63在免疫系统器官如轴静脉和前肾管中表达。综上所述,短期暴露于三唑酮,可能引起大型溞的氧化应激和神经毒性,而长期暴露则可能会引起免疫毒性,内分泌干扰作用将一直存在。
对比F1代和F2代的KEGG通路和关键基因,KEGG通路均与代谢和免疫相关,但F2代内分泌干扰作用和免疫反应更加明显,F1代更多表现在氧化应激和消化系统相关的通路。F1代上调基因占比高而F2代下调基因占比高,在F1代中显著上调的BDH相关基因在F2代中显著下调。这些差异可能是由于F1代的暴露降低了三唑酮对F2代的毒性阈值,从而加剧了F2代的毒性作用,长期暴露后,氧化损伤最终导致细胞凋亡和机体功能损失等。F1代在低浓度暴露时,生物体通过提高基因表达来适应污染环境,而高浓度时可能超过了部分基因表达毒性阈值从而引起基因表达量的变化。F2代在低浓度暴露时基因下调,可能是因为其母代F1暴露于三唑酮从而加剧了对子代的毒性。因此,随着时间和代际延长,三唑酮对大型溞的毒性作用可能会加强,大型溞对某一阈值浓度的三唑酮有一定的调控能力,但超过一定浓度,可能会存在传代效应。
综上所述,低浓度三唑酮长期暴露大型溞会引起慢性毒性,≥200 μg·L-1的三唑酮显著影响大型溞的体长和繁殖。三唑酮可能会引起大型溞的氧化应激反应、神经毒性、免疫毒性和内分泌干扰,存在传代效应,长期暴露会下调免疫反应等相关基因的表达,降低毒性阈值。
通讯作者简介:金小伟(1985—),男,博士,正高级工程师,主要研究方向为生态毒理学和生态风险评价。
[1] Watschke T L, Mumma R O, Linde D T, et al. Surface Runoff of Selected Pesticides Applied to Turfgrasses [M]// Fate and Management of Turfgrass Chemicals. Washington DC: American Chemical Society, 1999: 94-105
[2] 迭庆杞, 黄泽春, 杨玉飞, 等. 我国农药工业危险废物产生和污染特性研究[J]. 环境工程技术学报, 2021, 11(6): 1266-1272
Die Q Q, Huang Z C, Yang Y F, et al. Research on the generation and pollution characteristics of pesticide industrial hazardous wastes in China [J]. Journal of Environmental Engineering Technology, 2021, 11(6): 1266-1272 (in Chinese)
[3] Fu Y, Yang T, Zhao J, et al. Determination of eight pesticides in Lycium barbarum by LC-MS/MS and dietary risk assessment [J]. Food Chemistry, 2017, 218: 192-198
[4] 刘园, 杨卫萍, 魏琛, 等. 枯水期贵阳市饮用水源农药污染特征及健康风险[J]. 地球与环境, 2015, 43(6): 653-659
Liu Y, Yang W P, Wei C, et al. Pollution characteristics and health risk assessment of pesticide in drinking water of Guiyang City, China during withered water period [J]. Earth and Environment, 2015, 43(6): 653-659 (in Chinese)
[5] 刘娜, 金小伟, 薛荔栋, 等. 太湖流域药物和个人护理品污染调查与生态风险评估[J]. 中国环境科学, 2017, 37(9): 3515-3522
Liu N, Jin X W, Xue L D, et al. Concentrations distribution and ecological risk assessment of pharmaceuticals and personal care products in Taihu Lake [J]. China Environmental Science, 2017, 37(9): 3515-3522 (in Chinese)
[6] 游明华. 天然水中9种三唑类农药的检测方法及其非生物降解研究[D]. 厦门: 厦门大学, 2008: 39
You M H. Determination and abiotic degradation of nine trizole pesticides in natural aquatic environments [D]. Xiamen: Xiamen University, 2008: 39 (in Chinese)
[7] Wang Z K, Tian Z N, Chen L, et al. Stereoselective metabolism and potential adverse effects of chiral fungicide triadimenol on Eremias argus [J]. Environmental Science and Pollution Research International, 2020, 27(8): 7823-7834
[8] Xu P, Huang L D. Stereoselective bioaccumulation, transformation, and toxicity of triadimefon in Scenedesmus obliquus [J]. Chirality, 2017, 29(2): 61-69
[9] Liu T T, Diao J L, Di S S, et al. Stereoselective bioaccumulation and metabolite formation of triadimefon in Tubifex tubifex [J]. Environmental Science & Technology, 2014, 48(12): 6687-6693
[10] Li M, Li S Y, Yao T T, et al. Waterborne exposure to triadimefon causes thyroid endocrine disruption and developmental delay in Xenopus laevis tadpoles [J]. Aquatic Toxicology, 2016, 177: 190-197
[11] Zhang W J, Deng Y, Chen L, et al. Effect of triadimefon and its metabolite on adult amphibians Xenopus laevis [J]. Chemosphere, 2020, 243: 125288
[12] Zhang W J, Deng Y, Chen L, et al. Comparing the effect of triadimefon and its metabolite on male and female Xenopus laevis: Obstructed growth and gonad morphology [J]. Chemosphere, 2020, 259: 127415
[13] de la Paz J F, Beiza N, Paredes-Zú iga S, et al. Triazole fungicides inhibit zebrafish hatching by blocking the secretory function of hatching gland cells [J]. International Journal of Molecular Sciences, 2017, 18(4): 710
iga S, et al. Triazole fungicides inhibit zebrafish hatching by blocking the secretory function of hatching gland cells [J]. International Journal of Molecular Sciences, 2017, 18(4): 710
[14] Hassold E, Backhaus T. Chronic toxicity of five structurally diverse demethylase-inhibiting fungicides to the crustacean Daphnia magna: A comparative assessment [J]. Environmental Toxicology and Chemistry, 2009, 28(6): 1218-1226
[15] Paredes-Zú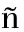 iga S, Trost N, de la Paz J F, et al. Behavioral effects of triadimefon in zebrafish are associated with alterations of the dopaminergic and serotonergic pathways [J]. Progress in Neuro-Psychopharmacology and Biological Psychiatry, 2019, 92: 118-126
iga S, Trost N, de la Paz J F, et al. Behavioral effects of triadimefon in zebrafish are associated with alterations of the dopaminergic and serotonergic pathways [J]. Progress in Neuro-Psychopharmacology and Biological Psychiatry, 2019, 92: 118-126
[16] Ward W O, Delker D A, Hester S D, et al. Transcriptional profiles in liver from mice treated with hepatotumorigenic and nonhepatotumorigenic triazole conazole fungicides: Propiconazole, triadimefon, and myclobutanil [J]. Toxicologic Pathology, 2006, 34(7): 863-878
[17] Liu H C, Chu T Y, Chen L L, et al. The cardiovascular toxicity of triadimefon in early life stage of zebrafish and potential implications to human health [J]. Environmental Pollution, 2017, 231: 1093-1103
[18] Jiang J H, Hu G J, Zhang C P, et al. Toxicological analysis of triadimefon on endocrine disruption and oxidative stress during rare minnow (Gobiocypris rarus) larvae development [J]. Environmental Science and Pollution Research International, 2017, 24(34): 26681-26691
[19] Wu S G, Hu G J, Zhao X P, et al. Synergistic potential of fenvalerate and triadimefon on endocrine disruption and oxidative stress during rare minnow embryo development [J]. Environmental Toxicology, 2018, 33(7): 759-769
[20] Zhang W J, Lu Y L, Huang L D, et al. Comparison of triadimefon and its metabolite on acute toxicity and chronic effects during the early development of Rana nigromaculata tadpoles [J]. Ecotoxicology and Environmental Safety, 2018, 156: 247-254
[21] Liu S Y, Jin Q, Huang X H, et al. Disruption of zebrafish (Danio rerio) sexual development after full life-cycle exposure to environmental levels of triadimefon [J]. Environmental Toxicology and Pharmacology, 2014, 37(1): 468-475
[22] 刘娜, 金小伟, 王业耀, 等. 三唑酮对青鳉鱼和大型溞不同测试终点的毒性效应评价[J]. 中国环境科学, 2016, 36(7): 2205-2211
Liu N, Jin X W, Wang Y Y, et al. Toxicity effect of triadimefon based on Oryzias latipes and Daphnia magna with different test endpoints [J]. China Environmental Science, 2016, 36(7): 2205-2211 (in Chinese)
[23] 刘娜, 金小伟, 穆云松, 等. 三唑酮在水环境中的环境行为、毒性效应及生态风险[J]. 生态毒理学报, 2017, 12(4): 65-75
Liu N, Jin X W, Mu Y S, et al. Review of environmental behavior, toxicity and ecological risk of triadimefon in the aquatic environment [J]. Asian Journal of Ecotoxicology, 2017, 12(4): 65-75 (in Chinese)
[24] 高嘉蔚, 赵莎莎, 李富云, 等. 微塑料对大型溞摄食和抗氧化防御系统的影响[J]. 环境科学研究, 2021, 34(5): 1205-1212
Gao J W, Zhao S S, Li F Y, et al. Effects of microplastics on feeding behavior and antioxidant system of Daphnia magna [J]. Research of Environmental Sciences, 2021, 34(5): 1205-1212 (in Chinese)
[25] Shaw J R, Pfrender M E, Eads B D, et al. Daphnia as an emerging model for toxicological genomics [J]. Advances in Experimental Biology, 2008, 2: 165-328
[26] Chen Y, Huang J, Xing L Q, et al. Effects of multigenerational exposures of D. magna to environmentally relevant concentrations of pentachlorophenol [J]. Environmental Science and Pollution Research, 2014, 21(1): 234-243
[27] Organization for Economic Co-operation and Development (OECD). Test No. 211: Daphnia magna reproduction test [R]. Paris: OECD, 2008
[28] Organization for Economic Co-operation and Development (OECD). Test No. 201: Freshwater alga and cyanobacteria: Growth inhibition [R]. Paris: OECD, 2008
[29] Quinn B, Gagné F, Blaise C, et al. Evaluation of the lethal and sub-lethal toxicity and potential endocrine disrupting effect of nonylphenol on the zebra mussel (Dreissena polymorpha) [J]. Comparative Biochemistry and Physiology Toxicology & Pharmacology, 2006, 142(1-2): 118-127
[30] Ford A T, LeBlanc G A. Endocrine disruption in invertebrates: A survey of research progress [J]. Environmental Science & Technology, 2020, 54(21): 13365-13369
[31] 胡方华, 宋文华, 丁峰, 等. 三唑酮对大型溞21天慢性毒性效应[J]. 生态毒理学报, 2012, 7(2): 171-176
Hu F H, Song W H, Ding F, et al. 21-d chronic toxicity of triadimefon to Daphnia magna [J]. Asian Journal of Ecotoxicology, 2012, 7(2): 171-176 (in Chinese)
[32] Cui F, Chai T T, Qian L, et al. Effects of three diamides (chlorantraniliprole, cyantraniliprole and flubendiamide) on life history, embryonic development and oxidative stress biomarkers of Daphnia magna [J]. Chemosphere, 2017, 169: 107-116
[33] Horwitz J. Alpha-crystallin [J]. Experimental Eye Research, 2003, 76(2): 145-153
[34] Walker Q D, Lewis M H, Crofton K M, et al. Triadimefon, a triazole fungicide, induces stereotyped behavior and alters monoamine metabolism in rats [J]. Toxicology and Applied Pharmacology, 1990, 102(3): 474-485
[35] Gagnaire F, Micillino J C. Effects of triadimefon on extracellular dopamine, DOPAC, HVA and 5-HIAA in adult rat striatum [J]. Toxicology, 2006, 217(2-3): 91-104
[36] Liu S Y, Chang J H, Zhao Y, et al. Changes of thyroid hormone levels and related gene expression in zebrafish on early life stage exposure to triadimefon [J]. Environmental Toxicology and Pharmacology, 2011, 32(3): 472-477
[37] Kenneke J F, Mazur C S, Ritger S E, et al. Mechanistic investigation of the noncytochrome P450-mediated metabolism of triadimefon to triadimenol in hepatic microsomes [J]. Chemical Research in Toxicology, 2008, 21(10): 1997-2004
[38] Jones E L, Demaria M C, Wright M D. Tetraspanins in cellular immunity [J]. Biochemical Society Transactions, 2011, 39(2): 506-511
[39] Termini C M, Gillette J M. Tetraspanins function as regulators of cellular signaling [J]. Frontiers in Cell and Developmental Biology, 2017, 5: 34
[40] Jung S, Kim M J, Sellaththurai S, et al. Generation of Cd63-deficient zebrafish to analyze the role of Cd63 in viral infection [J]. Fish & Shellfish Immunology, 2021, 111: 152-159