抗生素耐药性问题日益严重,对生态安全与公共健康构成重大威胁,预计到2050年,抗生素耐药性问题将导致全球每年1 000万人的死亡,比癌症导致的全球死亡人数多180万[1]。四环素类抗生素是一类广谱类抗生素,主要通过阻止菌体蛋白的合成对细菌产生作用,广泛应用于临床治疗、畜牧和水产养殖业[2]。据统计,四环素类抗生素每年全球的产量为数千吨[3],在全球的抗生素生产量和使用量中排名第二[4]。2010—2015年,在全球兽用抗生素的使用中,四环素类抗生素使用量位列第一,占全球兽用抗生素使用量的48%[5]。近几十年来,四环素、土霉素、金霉素和多西环素等一代、二代四环素类药物是世界上最常用的抗生素[4]。
四环素类抗生素耐药机制已被广泛报道,其耐药基因分布于约130种革兰氏阳性菌属和革兰氏阴性菌属并广泛存在于人类、动物和环境中[2,6]。四环素类抗生素的耐药机制包括外排泵[7-9]、核糖体保护蛋白[10-13]和酶失活机制[14]。外排泵将四环素类抗生素泵出胞外从而降低了其胞内浓度,核糖体保护蛋白与四环素类抗生素竞争在核糖体30S亚基的结合位点,酶失活则是产生氧化还原酶使四环素失活。外排泵和核糖体保护蛋白是临床中对四环素类药物最常见的耐药类型[15]。自从第一批四环素类药物被发现以来,核心结构的不断优化使四环素类药物在临床应用和开发中能够克服许多耐药性机制[15]。然而,新的四环素类抗生素耐药基因正不断增加,在全世界范围内快速传播,严重影响了抗菌药物治疗的有效性。
替加环素是首个甘氨酰环素类半合成抗生素[16],其属于第三代新型四环素类药物,具有超广谱的抗菌活性,用于治疗由多重耐药病原菌引起的感染性疾病[17]。世界卫生组织(WHO)于2019年将替加环素列为极其重要的抗菌药物[18]。然而,有研究人员已经从动物和人体内发现了质粒介导的高水平替加环素耐药基因tet(X)变异体——tet(X3)和tet(X4)[5]。tet(X3)和tet(X4)不仅可以引起细菌对替加环素的高水平耐药,最小抑菌浓度(minimum inhibitory concentration, MIC)为32~64 μg·mL-1,还可介导2个第四代四环素类新药奥马环素和依拉环素的高水平耐药。值得注意的是,新型tet(X)变异体还存在交叉耐药性,可介导对传统的一代、二代四环素类药物耐药。我国不同养殖动物和环境样品中均发现了携带不同tet(X)变异体的细菌[19-22]。2021年,我国学者从来自世界四大洲(亚洲、欧洲、北美洲和南美洲)的12 829个人类微生物组样本中鉴定出322个tet(X)变异体阳性样本[23]。因此,四环素类抗生素尤其是替加环素的耐药机制和流行传播迫切需要全球关注。本文综述了四环素类抗生素耐药机制研究进展,报道了新的四环素类抗生素耐药基因;重点阐述了质粒介导的替加环素耐药机制,详述了tet(X)基因及其变异基因的产生与发展;利用生物信息学分析了大量公共数据库已有细菌基因组,解析了替加环素耐药基因的细菌宿主以及tet(X)基因家族在细菌染色体和质粒中的分布特征,为进一步监测替加环素耐药基因的传播扩散提供科学参考。
1 四环素类抗生素作用机制与耐药机制(Action mechanisms and resistance mechanisms of tetracyclines)
1.1 四环素类抗生素化学结构特征
四环素类抗生素发现于20世纪40年代,目前已经发展到第四代[2]。四环素类抗生素为广谱抗生素,其名称来源于4个芳香环的结合,并四苯四环素核是所有四环素类抗生素共同的基本结构。金霉素(1948年)、土霉素(1950年)和四环素(1953年)是四环素家族的第一代成员(图1);第二代以米诺环素(1961年)和多西环素(1967年)为代表;替加环素(1993年)于2005年获得美国食品药品监督管理局(FDA)批准用于临床治疗,是四环素类唯一的第三代成员;奥马环素(2013年)和依拉环素(2013年)属于第四代成员,均于2018年获得FDA批准用于临床治疗。目前,替加环素、奥马环素和依拉环素属于新型四环素类抗生素。
替加环素是一种化学修饰的米诺环素,其核心D环C9处的氢被N, N-二甲基甘氨酰胺取代,从而赋予替加环素独特的微生物学特性和组织穿透性,替加环素抗菌谱得到扩展,其抑制蛋白质合成的能力是米诺环素的3倍,四环素的20倍,且不受四环素类抗生素主要耐药机制的影响[24],因此,替加环素可以治疗多重耐药病原体引起的感染,包括耐甲氧西林金黄色葡萄球菌(methicillin resistant Staphylocous aureus, MRSA)、耐青霉素肺炎链球菌(penicillin resistant Streptococcus pneumoniae, PRSP)、耐万古霉素肠球菌(vancomycin resistant Enterococcus, VRE)以及产超广谱β-内酰胺酶(extended spectrum beta-lactamases, ESBLs)大肠杆菌和肺炎克雷伯菌[25]。奥马环素是氨甲基四环素类药物,在D环的C7处有一个二甲氨基,在D环C9处有一个氨甲基修饰,C7官能团和C9的修饰保障了奥马环素对抗外排泵和核糖体保护蛋白耐药机制的稳定性[26]。依拉环素是含氟四环素,结构类似于替加环素,其对革兰氏阳性球菌的抗菌能力是替加环素的2倍~4倍,对革兰氏阴性杆菌的抗菌能力是替加环素的2倍~8倍[27]。
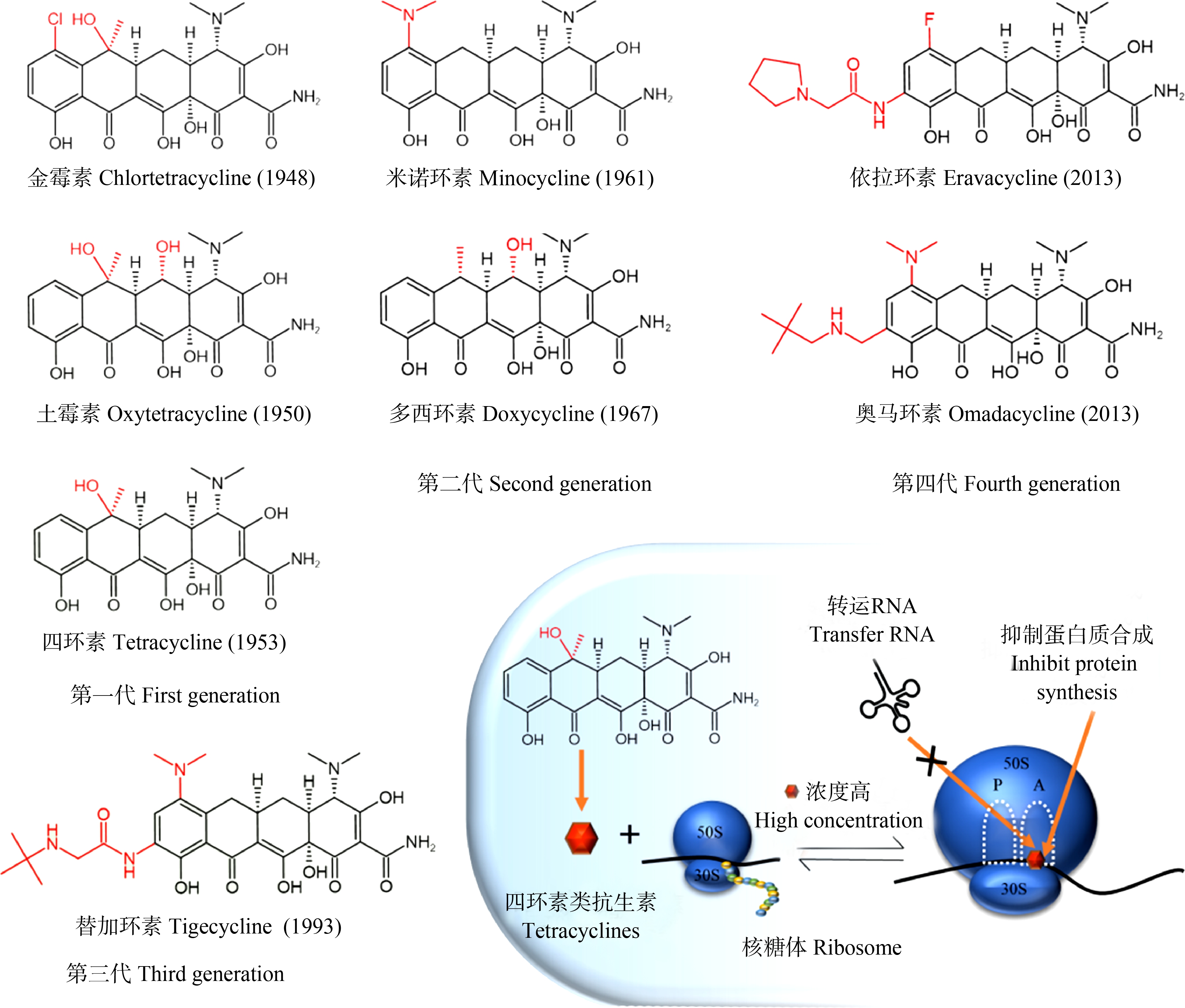
图1 典型四环素类抗生素的化学结构
Fig. 1 Chemical structures of representative tetracyclines
四环素类抗生素可逆地与细菌核糖体30S亚基结合,阻止了氨酰tRNA与核糖体结合位点A的结合,从而抑制细菌蛋白的翻译过程(图1),由于这种结合的可逆性,四环素类抗生素在细菌体内浓度较低时会从作用靶点离开。四环素抗菌作用能力强,而对人类副作用小的原因在于四环素与真核细胞核糖体80S亚基的结合能力相对较弱[28]。
1.2 四环素类抗生素耐药机制
细菌对四环素类抗生素耐药的分子机制包括4种:四环素类特异性外排泵、核糖体保护、酶失活和多药外排泵(图2)。根据华盛顿大学更新的一个四环素类耐药基因数据库(http://faculty.washington.edu/marilynr/; updated April 20, 2021),四环素类特异性外排泵基因共有38个,核糖体保护基因共13个,酶修饰基因共13个,未知分类基因1个,镶嵌式核糖体保护基因(mosiac ribosomal protection)共11个,四环素类耐药基因在GenBank中的代表序列登录号以及基因位置如表1所示。
细菌主动外排泵系统分为五大家族:主要易化超家族(major facilitator super family, MFS)、小多重耐药家族(small multidrug resistance, SMR)、ATP结合盒家族(ATP binding cassette, ABC)、耐药结节分化家族(resistance nodulation and cell division, RND)、多药和毒性化合物外排家族(multidrug and toxic compound extrusion family, MATE)。MFS、SMR和RND类外排泵均以质子驱动力为能量将药物泵出膜外,ABC类外排泵利用ATP水解释放的能量将抗生素泵出膜外[29-31]。38个四环素类特异性外排泵属于MFS家族,已发现的可外排四环素类抗生素的多药外排泵有RND、MFS和ABC家族[31]。RND家族四环素类多药外排泵主要有AdeABC、AdeIJK、AcrAB-TolC、MexAB-OprM和MexVW-OprM等;MFS家族四环素类多药外排泵主要有MdfA、NorA、NorB、Rv1410c和Rv2333c等;ABC家族四环素类多药外排泵主要有Rv1258C、EfrAB和VcaM等[31-33]。

图2 四环素类抗生素耐药机制
注:NADPH表示还原型烟酰胺腺嘌呤二核苷酸磷酸,NADP+表示烟酰胺腺嘌呤二核苷酸磷酸,MFS表示主要易化超家族,ABC表示ATP结合盒家族,RND表示耐药结节分化家族。
Fig. 2 Mechanisms of tetracycline resistance
Note: NADPH stands for reduced nicotinamide adenine dinucleotide phosphate, NADP+ stands for nicotinamide adenine dinucleotide phosphate, MFS stands for major facilitator super family, ABC stands for ATP binding cassette, and RND stands for resistance nodulation and cell division.
表1 四环素类抗生素耐药基因类型及其来源
Table 1 Types and sources of tetracycline resistance genes
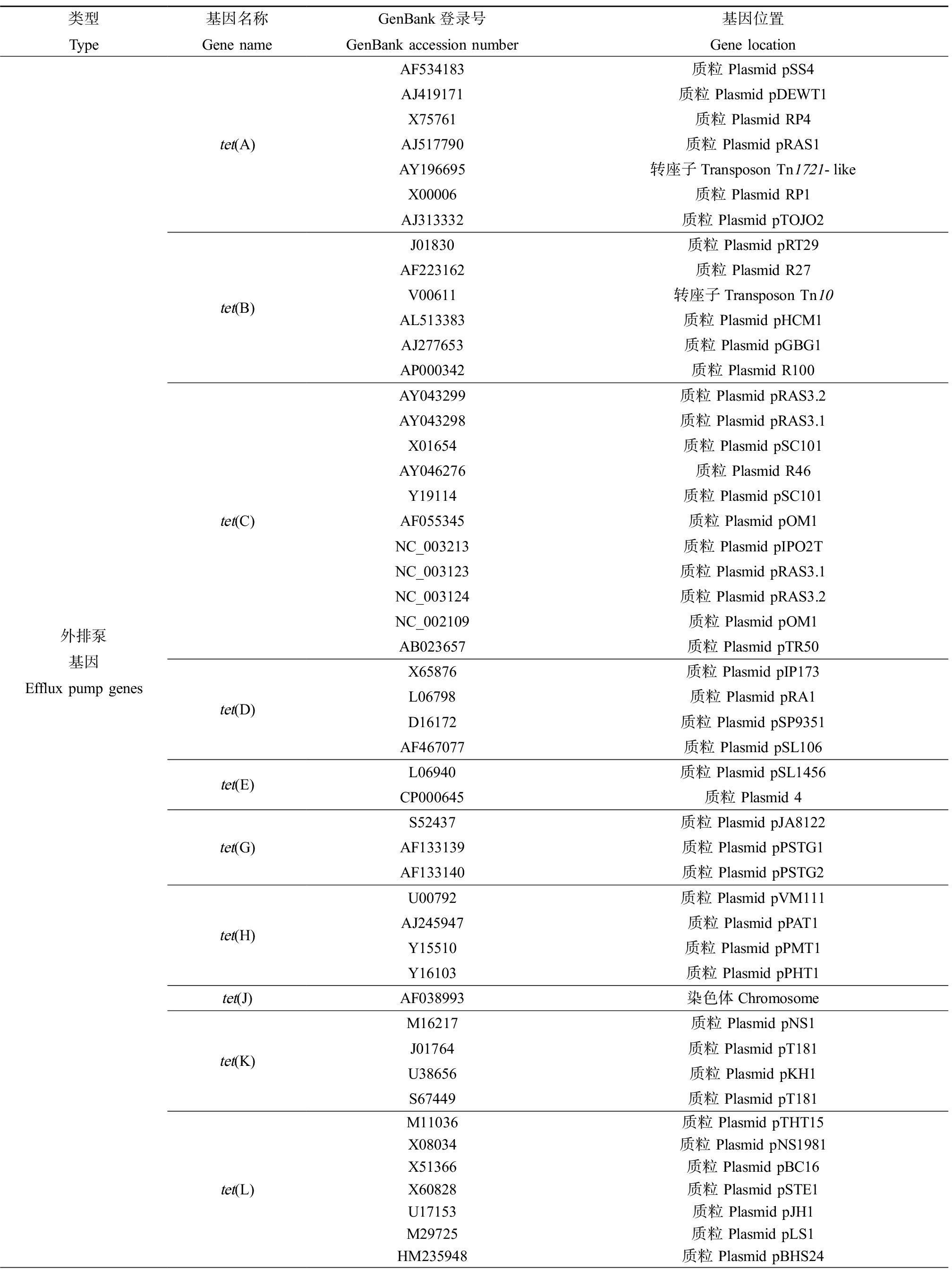
类型Type基因名称Gene nameGenBank登录号GenBank accession number基因位置Gene location外排泵基因Efflux pump genestet(A)tet(B)tet(C)tet(D)tet(E)tet(G)tet(H)tet(J)tet(K)tet(L)AF534183质粒 Plasmid pSS4AJ419171质粒 Plasmid pDEWT1X75761质粒 Plasmid RP4AJ517790质粒 Plasmid pRAS1AY196695转座子Transposon Tn1721-likeX00006质粒 Plasmid RP1AJ313332质粒 Plasmid pTOJO2J01830质粒 Plasmid pRT29AF223162 质粒 Plasmid R27V00611转座子Transposon Tn10AL513383质粒 Plasmid pHCM1AJ277653质粒 Plasmid pGBG1AP000342质粒 Plasmid R100AY043299质粒 Plasmid pRAS3.2AY043298质粒 Plasmid pRAS3.1X01654质粒 Plasmid pSC101AY046276质粒 Plasmid R46Y19114质粒 Plasmid pSC101AF055345 质粒 Plasmid pOM1NC_003213质粒 Plasmid pIPO2TNC_003123 质粒 Plasmid pRAS3.1NC_003124 质粒 Plasmid pRAS3.2NC_002109质粒 Plasmid pOM1AB023657质粒 Plasmid pTR50X65876质粒 Plasmid pIP173L06798质粒 Plasmid pRA1D16172 质粒 Plasmid pSP9351AF467077质粒 Plasmid pSL106L06940质粒 Plasmid pSL1456CP000645质粒 Plasmid 4S52437质粒 Plasmid pJA8122AF133139质粒 Plasmid pPSTG1AF133140质粒 Plasmid pPSTG2U00792质粒 Plasmid pVM111AJ245947质粒 Plasmid pPAT1Y15510质粒 Plasmid pPMT1Y16103质粒 Plasmid pPHT1AF038993染色体ChromosomeM16217质粒 Plasmid pNS1J01764质粒 Plasmid pT181U38656质粒 Plasmid pKH1S67449质粒 Plasmid pT181M11036质粒 Plasmid pTHT15X08034质粒 Plasmid pNS1981X51366 质粒 Plasmid pBC16X60828质粒 Plasmid pSTE1U17153质粒 Plasmid pJH1M29725质粒 Plasmid pLS1HM235948质粒 Plasmid pBHS24
续表1
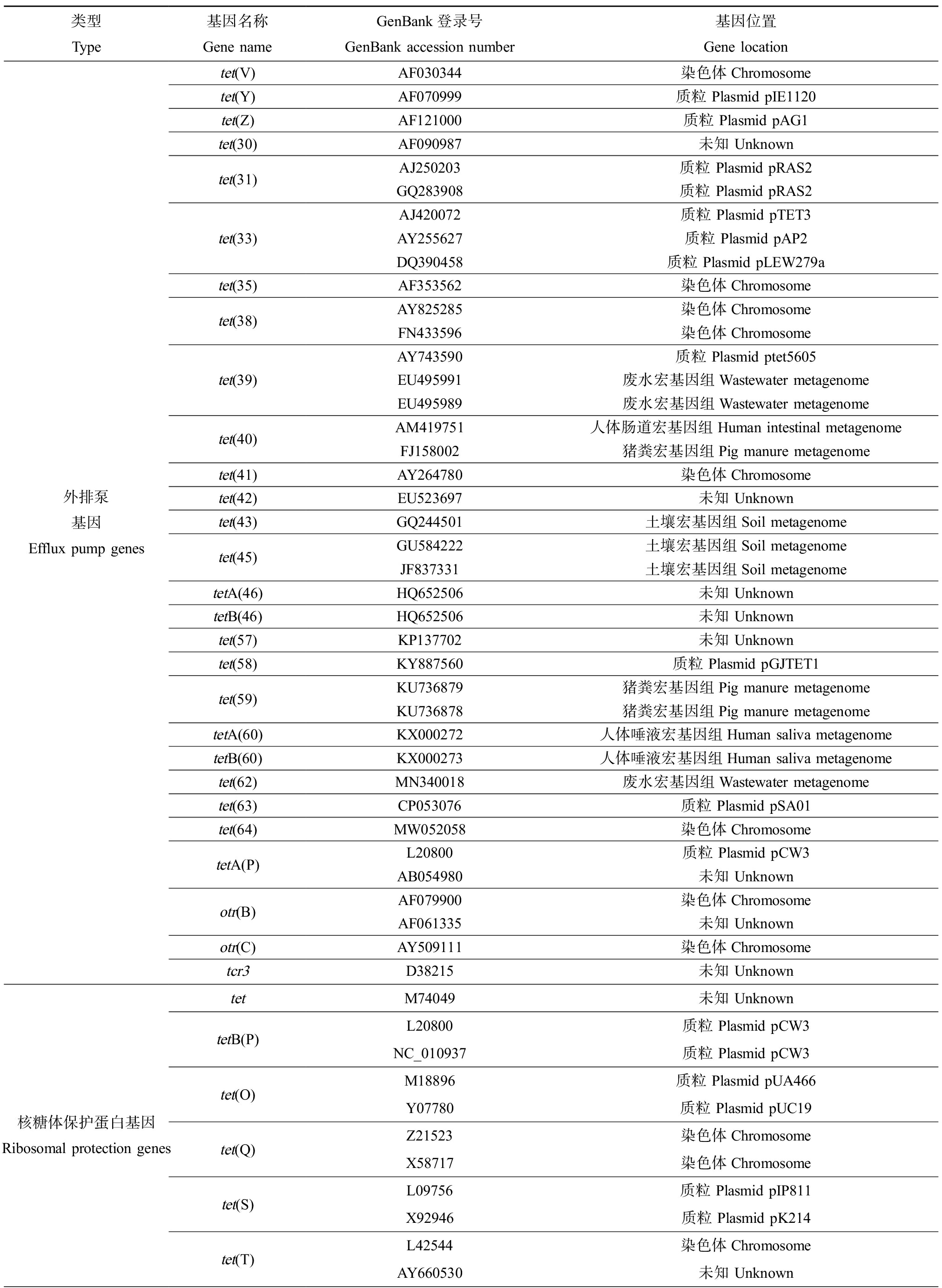
类型Type基因名称Gene nameGenBank登录号GenBank accession number基因位置Gene location外排泵基因Efflux pump genestet(V)tet(Y)tet(Z)tet(30)tet(31)tet(33)tet(35)tet(38)tet(39)tet(40)tet(41)tet(42)tet(43)tet(45)tetA(46)tetB(46)tet(57)tet(58)tet(59)tetA(60)tetB(60)tet(62)tet(63)tet(64)tetA(P)otr(B)otr(C)tcr3AF030344染色体ChromosomeAF070999质粒 Plasmid pIE1120AF121000质粒 Plasmid pAG1AF090987未知 UnknownAJ250203质粒 Plasmid pRAS2GQ283908质粒 Plasmid pRAS2AJ420072质粒 Plasmid pTET3AY255627 质粒 Plasmid pAP2DQ390458质粒 Plasmid pLEW279aAF353562染色体ChromosomeAY825285染色体ChromosomeFN433596染色体ChromosomeAY743590质粒 Plasmid ptet5605EU495991废水宏基因组Wastewater metagenomeEU495989废水宏基因组Wastewater metagenomeAM419751人体肠道宏基因组Human intestinal metagenomeFJ158002猪粪宏基因组Pig manure metagenomeAY264780染色体ChromosomeEU523697未知 UnknownGQ244501土壤宏基因组Soil metagenomeGU584222 土壤宏基因组Soil metagenomeJF837331土壤宏基因组Soil metagenomeHQ652506未知 UnknownHQ652506未知 UnknownKP137702未知 UnknownKY887560质粒 Plasmid pGJTET1KU736879猪粪宏基因组Pig manure metagenomeKU736878猪粪宏基因组Pig manure metagenomeKX000272人体唾液宏基因组Human saliva metagenomeKX000273人体唾液宏基因组Human saliva metagenomeMN340018废水宏基因组Wastewater metagenomeCP053076质粒 Plasmid pSA01MW052058染色体ChromosomeL20800质粒 Plasmid pCW3AB054980未知 UnknownAF079900染色体ChromosomeAF061335未知 UnknownAY509111染色体ChromosomeD38215未知 Unknown核糖体保护蛋白基因Ribosomal protection genestettetB(P)tet(O)tet(Q)tet(S)tet(T)M74049未知 UnknownL20800质粒 Plasmid pCW3NC_010937质粒 Plasmid pCW3M18896质粒 Plasmid pUA466Y07780质粒 Plasmid pUC19Z21523染色体ChromosomeX58717染色体ChromosomeL09756质粒 Plasmid pIP811X92946质粒 Plasmid pK214L42544染色体ChromosomeAY660530未知 Unknown
续表1
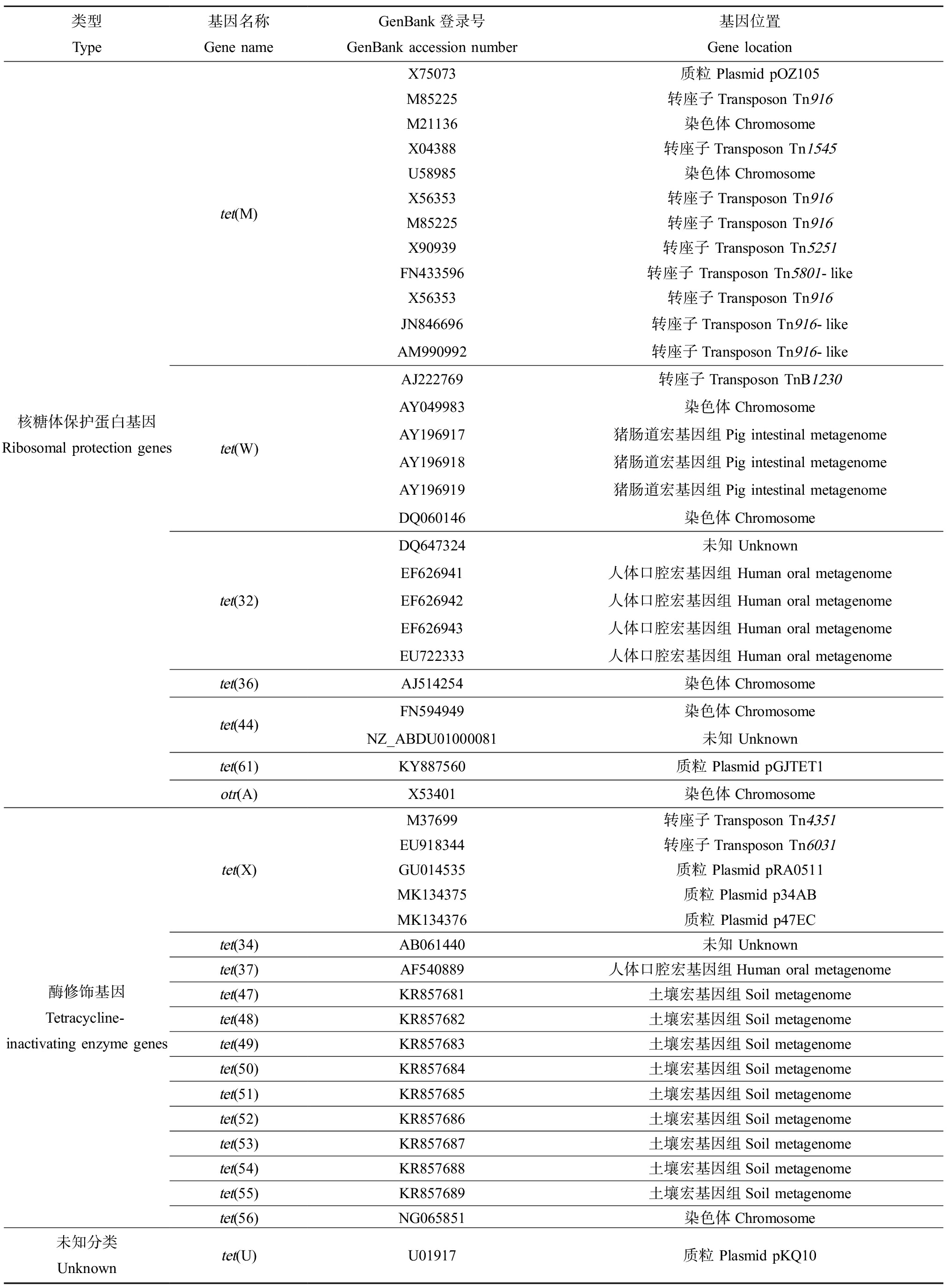
类型Type基因名称Gene nameGenBank登录号GenBank accession number基因位置Gene location核糖体保护蛋白基因Ribosomal protection genestet(M)tet(W)tet(32)tet(36)tet(44)tet(61)otr(A)X75073质粒 Plasmid pOZ105M85225 转座子Transposon Tn916M21136染色体ChromosomeX04388转座子Transposon Tn1545U58985染色体ChromosomeX56353转座子Transposon Tn916M85225转座子Transposon Tn916X90939转座子 Transposon Tn5251FN433596转座子 Transposon Tn5801-likeX56353 转座子Transposon Tn916JN846696转座子Transposon Tn916-likeAM990992转座子Transposon Tn916-likeAJ222769转座子Transposon TnB1230AY049983染色体ChromosomeAY196917 猪肠道宏基因组Pig intestinal metagenomeAY196918猪肠道宏基因组Pig intestinal metagenomeAY196919猪肠道宏基因组Pig intestinal metagenomeDQ060146染色体ChromosomeDQ647324未知 UnknownEF626941 人体口腔宏基因组 Human oral metagenomeEF626942人体口腔宏基因组 Human oral metagenomeEF626943人体口腔宏基因组 Human oral metagenomeEU722333人体口腔宏基因组 Human oral metagenomeAJ514254染色体ChromosomeFN594949染色体ChromosomeNZ_ABDU01000081未知 UnknownKY887560质粒 Plasmid pGJTET1X53401染色体Chromosome酶修饰基因Tetracycline-inactivating enzyme genestet(X)tet(34)tet(37)tet(47)tet(48)tet(49)tet(50)tet(51)tet(52)tet(53)tet(54)tet(55)tet(56)M37699转座子Transposon Tn4351EU918344转座子Transposon Tn6031GU014535质粒 Plasmid pRA0511MK134375质粒 Plasmid p34ABMK134376质粒 Plasmid p47ECAB061440未知 UnknownAF540889人体口腔宏基因组Human oral metagenomeKR857681土壤宏基因组Soil metagenomeKR857682土壤宏基因组Soil metagenomeKR857683土壤宏基因组Soil metagenomeKR857684土壤宏基因组Soil metagenomeKR857685土壤宏基因组Soil metagenomeKR857686土壤宏基因组Soil metagenomeKR857687土壤宏基因组Soil metagenomeKR857688土壤宏基因组Soil metagenomeKR857689土壤宏基因组Soil metagenomeNG065851染色体Chromosome未知分类Unknowntet(U)U01917质粒 Plasmid pKQ10
四环素类抗生素耐药性通常是通过水平转移获得的[34-35]。2015年,Forsberg等[35]从土壤宏基因组中发现了9个通过酶失活机制导致高水平四环素抗性的基因,命名为tet(47)~tet(55),其中tet(47)和tet(54)与氨基糖苷磷酸转移酶耐药基因相邻。tet(62)、tet(63)和tet(64)为最新发现的四环素类抗生素耐药基因[36-38]。McGivern等[36]在暴露于抗菌剂三氯生的合成废水中识别出了新的外排泵基因tet(62),将携带tet(62)的基因文库导入大肠杆菌后,抗生素敏感性分析表明细菌对所测试的四环素类抗生素耐药性增加了4倍~50倍。Zhu等[37]在来源于鸡样本的金黄色葡萄球菌多重耐药质粒pSA01(25 664 bp)上发现一个新的外排泵基因tet(63),该质粒同时携带了aadD、aacA-aphD和erm(C)等耐药基因,Tet(63)蛋白与外排泵蛋白Tet(K)的同源性为73%。Somprasong等[38]在来源于土壤样本的乌邦伯克霍尔德菌(Burkholderia ubonensis)中发现新的外排泵基因tet(64),对四环素呈现高水平耐药性(MIC≥256 μg·mL-1),但不能介导米诺环素、替加环素和依拉环素抗性。
2 替加环素耐药机制(Mechanisms of tigecycline resistance)
替加环素是一种新型甘氨酰环肽类抗生素,属于第三代四环素类抗生素,与核糖体30S亚基的结合效率是四环素的5倍[39]。替加环素不受四环素类抗生素主要耐药机制的影响,如四环素特异性外排泵和核糖体保护机制;也不受β内酰胺酶、靶位修饰、大环内酯类外排泵或酶靶位改变(如旋转酶/拓扑异构酶)等耐药机制的影响。这些特性使得替加环素具有超广谱抗菌活性,是治疗产碳青霉烯酶多重耐药肠杆菌感染的“最后一道防线”[17]。目前,替加环素已被批准作为一种单药治疗复杂感染[40]。Hoban等[41]于2004年开展了替加环素评估和监测试验,评估了替加环素和常用抗生素对来自11个国家40个研究中心的6 792个临床分离株的抗菌活性,其药敏实验结果表明,替加环素对粪肠球菌、屎肠球菌、金黄色葡萄球菌、无乳链球菌和肺炎链球菌的MIC90分别为0.12、0.12、0.25和0.25 μg·mL-1,对肠杆菌科细菌MIC90为1 μg·mL-1。因此,替加环素的重要性体现在它对大多数多重耐药菌具有抗菌活性,被世界卫生组织列为极其重要的抗菌药物[5]。
随着替加环素使用量的增加,现有研究表明替加环素耐药性已经出现,临床上最常见于鲍曼不动杆菌和肠杆菌科,尤其是多药耐药菌株,替加环素耐药机制的产生与发展已得到广泛关注[5,42-44]。目前发现的替加环素耐药机制有:核糖体蛋白结合位点突变、细胞膜变异、主动外排泵转运系统和修饰酶降解等。其中核糖体蛋白结合位点突变和细胞膜变异的替加环素耐药机制报道数量较少:在肺炎克雷伯菌、粪肠球菌、鲍曼不动杆菌、大肠杆菌和金黄色葡萄球菌中,核糖体S10蛋白的编码基因rpsJ突变可介导替加环素耐药[45-47];plsC基因突变和abrp基因缺失导致鲍曼不动杆菌细胞膜通透性改变,从而介导替加环素耐药[48-49];2021年,我国学者在猪链球菌中发现了对替加环素具有抗性的核糖体保护蛋白Tet(M)的突变体,其编码基因tet(M)位于基因岛上[50]。鉴于替加环素在临床治疗中的重要性,有关其耐药机制的报道正不断增加,本文将详述替加环素2种主要耐药机制外排泵和修饰酶的形成与发展(图3)。
2.1 替加环素外排泵
与替加环素耐药相关的外排泵主要有RND家族、MFS家族中的Tet(A)和Tet(L)外排泵突变体(图3)。
2.1.1 RND家族外排泵
RND家族外排泵常见于革兰氏阴性菌,如铜绿假单胞菌、奇异变形杆菌和摩氏摩根菌[43]。RND外排泵主要利用质子梯度来驱动外排,1个H+交换1个药物分子。与替加环素耐药相关的RND型外排泵包括:AcrAB-TolC[51]、SdeXY-HasF[52]、OqxAB[43,53]、TMEXcd1-TOprJ[54]和AdeABC/AdeIJK/AdeFGH[55]。
在大肠杆菌中,RND家族外排泵AcrAB-TolC由三联组合蛋白组成,包括膜连接蛋白(AcrA)、内膜转运蛋白(AcrB)和外膜通道蛋白(TolC)(图3)。AcrB捕获和转运底物,在AcrA的协助下,激活外排泵通道,将底物泵出细菌胞外。该外排泵的调控主要有局部调控(AcrR)和全局调控(MarA、SoxS、Rob和RamA)(图3)。acrA和acrB基因由同一个操纵子转录而来,AcrR是一种负调控因子,调控acrAB的表达[51]。在肠杆菌科细菌中,AcrAB外排泵是替加环素的主要耐药机制,其局部和全局调控因子则因物种而异[42]。在大肠杆菌中,当替加环素进入细菌内,局部调控蛋白AcrR与之结合引发构象改变,从而激活外排蛋白AcrAB的过表达,导致大肠杆菌对替加环素的敏感性降低[56]。在AcrAB-TolC外排泵全局调控因子中,现有研究表明ramA表达增强促成了阴沟肠杆菌、产气肠杆菌和克雷伯氏菌的替加环素耐药性[53,57]。

图3 替加环素主要耐药机制
Fig. 3 Main mechanisms of tigecycline resistance
OqxAB外排泵是RND家族中第一个由质粒介导的外排泵,由膜融合蛋白OqxA和内膜转运蛋白OqxB组成。Veleba等[58]对肺炎克雷伯菌的研究结果表明,rarA的过表引起OqxAB和AcrAB表达上调,进而导致包括替加环素在内的多耐药表型。此外,该项研究还发现OqxAB外排泵受到OqxR的负调控。Veleba等[53]在另一项研究中发现阴沟肠杆菌和产气肠杆菌对替加环素耐药的主要介导因素为RamR调控引起的RamA表达增加。随后,Jiménez-Castellanos等[59]对肺炎克雷伯菌的研究证实了OqxAB同时受RamA和RarA的调控。综上所述,OqxAB同时受到RarA、RamA以及OqxR的调控,其中RarA和RamA调控蛋白介导细菌对替加环素的耐药性。
SdeXY-HasF外排泵系统由膜融合蛋白(SdeX)、内膜转运蛋白(SdeY)和外膜蛋白(HasF)组成。SdeXY是在机会致病菌——粘质沙雷氏菌(Serratia marcescens)中发现的第一个RND型外排泵,是一种能量依赖的药物外排泵,具有广泛的药物底物,在粘质沙雷氏菌多重耐药过程中发挥了重要作用[60]。HasF是粘质沙雷菌的TolC同系物,参与能量依赖的流出[61]。SdeXY-HasF外排泵系统的上调与粘质沙雷氏菌对替加环素的耐药性有关,进一步的研究需要阐明SdeXY过表达的机制[52]。
TMexCD1-TOprJ1是最近在动物源克雷伯氏菌中发现的由IncFIA质粒编码的RND型外排泵[54]。TMexCD1-TOprJ1与铜绿假单胞菌染色体编码的MexCD-OprJ外排泵氨基酸同源性为64.5%~77.8%,在大肠杆菌中表达TMexCD1-TOprJ1导致替加环素外排增加。tmexcd1-toprj1基因在动物源肠杆菌和假单胞菌中较为常见[62],然而近2年在人医临床分离的细菌中陆续有报道该基因的检出[63-64],TMexCD1-TOprJ1的肠杆菌已扩散到食品市场的环境污水[65]。目前,已鉴定出其他2种类似的外排泵,其中TMexCD2-TOprJ2位于解鸟氨酸拉乌尔菌(Raoultella ornithinolytica)的质粒和染色体[66],TMexCD3-TOprJ3发现于动物源变形杆菌染色体(Proteus mirabilis)[67]。这一类型外排泵基因主要分布在克雷伯氏菌的杂合质粒中,如IncHI1B-FIB质粒、IncC-IncX3质粒[62,65,68]。
AdeABC、AdeIJK和AdeFGH这3种RND家族外排泵被认为与鲍曼不动杆菌对替加环素的耐药有关[55,69-70],其中,AdeABC外排泵系统在临床分离的产碳青霉烯酶鲍曼不动杆菌中最为常见[71]。该外排泵系统由膜连接蛋白AdeA、内膜转运蛋白AdeB和外膜通道蛋白AdeC组成,其转录水平受到AdeRS双组分系统的调控。
2.1.2 MFS家族外排泵
Tet(A)外排泵,是临床上发现的最常见的四环素MFS型外排泵[72],主要存在于革兰氏阴性菌中。Tet(A)蛋白具有12个跨膜α-螺旋片段,其表达受到TetR蛋白的转录调控,TetR蛋白属于阻遏蛋白,四环素通过结合TetR阻遏蛋白将其从操纵子O1、O2上解离下来,进而促进tet(A)的表达[73]。虽然替加环素克服了四环素特异性外排泵Tet(A)的耐药机制,但tet(A)突变体能介导细菌对替加环素高水平耐药[52]。Hentschke等[74]对一株呈现替加环素高水平耐药(MIC=16 μg·mL-1)的沙门菌进行研究,分子机制分析表明tet(A)突变和ramR失活介导沙门菌对替加环素的高水平耐药,且tet(A)突变体位于质粒上的转座子Tn1721中。质粒携带的tet(A)基因在临床上产碳青霉烯酶肺炎克雷伯菌中广泛存在,tet(A)的突变是导致替加环素耐药性的主要原因[75-76]。
Tet(L)是MFS家族外排泵中一种具有14个跨膜片段的四环素外排泵蛋白[77-78],主要存在于革兰氏阳性菌中。Tet(L)蛋白与Tet(K)类似,都能催化四环素-二价金属离子Me2+复合物与H+逆向转运而介导四环素类抗生素耐药(图3)[79]。tet(L)基因于1992年首次在葡萄球菌的质粒pSTE1中报道[80],常在革兰氏阳性菌(如芽孢杆菌、葡萄球菌、链球菌和肠球菌)的质粒中被发现,通常位于大小、结构不同的质粒上,并与四环素类耐药基因tet(T)和tet(K)、氨基糖苷类耐药基因aadD共存[81-82]。在革兰氏阴性菌(如巴氏杆菌和曼海姆氏菌)的质粒中偶尔也有报道[83]。Tet(L)外排泵可以输出四环素和强力霉素,但不能外排替加环素。然而,已有研究表明,Tet(L)外排泵过表达可导致替加环素耐药,如在肠球菌中,Tet(L)外排泵和Tet(M)核糖体保护蛋白的过表达介导了替加环素的耐药[84]。此外,Tet(L)外排泵突变也能介导替加环素耐药,我国报道了新型Tet(L)外排泵变异体——tet(L)F58L和tet(L)A117V,可介导葡萄球菌、弯曲杆菌对替加环素和依拉环素的耐药性[85-86]。
2.2 替加环素耐药基因tet(X)及其变异体的产生与流行
替加环素修饰酶是由tet(X)及其变异基因编码的黄素依赖性单加氧酶。Tet(X)由388个氨基酸组成,在黄素腺嘌呤二核苷酸(FAD)、还原型烟酰胺腺嘌呤二核苷酸磷酸(NADPH)、氧气(O2)和镁离子(Mg2+)存在的条件下具有催化活性,可催化降解传统以及新型四环素类抗生素,pH为8.5时该酶的活性最高[14]。到目前为止,质粒携带的可转移tet(X)及其变异体有tet(X)、tet(X3)、tet(X4)、tet(X5)、tet(X6)和tet(X7),可介导对替加环素的高水平耐药,其编码的Tet(X)修饰酶可羟化替加环素的C11a,生成11a-羟基替加环素(图3)。tet(X)基因家族在不同来源细菌或环境中的流行情况如表2所示。
2.2.1 tet(X)
tet(X)是第一个能直接灭活四环素的抗性基因。1982年,Tally等[87]在拟杆菌质粒pBFTM10上发现新的四环素耐药基因。1984年,Guiney等[88]发现位于质粒pBF4的类似的耐药基因,研究人员将该基因克隆到大肠杆菌载体中,药敏试验结果表明转化菌株在好氧条件下表达四环素耐药性,而在拟杆菌属(严格的厌氧菌)中不表达四环素耐药性。1986年,Matthews和Guiney[89]的研究表明新的四环素耐药基因位于质粒pBFTM10上的转座子Tn4400和质粒pBF4上的转座子Tn4351。1989年,Speer和Salyers[90]证明了这种四环素抗性基因产物对四环素进行了化学修饰,该基因是第一个通过化学修饰四环素的耐药机制的例子。1990年,Speer和Salyers[91]认为该四环素基因不与已知的四环素耐药基因杂交,属于单独的一类耐药基因,并将该基因命名为tet(X)。
2.2.2 tet(X1)、tet(X2)
2001年,Whittle等[92]首次在拟杆菌属结合转座子CTnDOT上发现了tet(X1)基因和tet(X2)基因,tet(X2)编码的蛋白与tet(X)编码的蛋白有99%的氨基酸同源性,然而tet(X1)编码的蛋白与tet(X)编码的蛋白仅有66%的氨基酸同源性。从tet(X)及其变体的首次发现至今,tet(X)/tet(X2)基因已在全球范围内广泛传播。2020年,Fang等[2]的流行病学回溯性研究表明,tet(X)/tet(X2)基因传播范围已超过16个国家/地区,覆盖七大洲中的五大洲,并广泛存在于各种生态系统中,包括人体肠道、土壤、沉积物、医院废水、污水厂污水、河水、牲畜粪便和环境气溶胶。
2.2.3 tet(X3)、tet(X3.2)、tet(X4)
He等[5]于2019年首次发现了质粒介导的高水平替加环素耐药基因tet(X3)和tet(X4),分别位于p34AB质粒和p47EC质粒上。Tet(X3)和Tet(X4)灭活所有的四环素家族,包括替加环素和FDA新批准的依拉环素和奥马环素。2019年,tet(X3.2)首次从来源于虾样本的短稳杆菌(Empedobacter brevis)中鉴定出,且位于质粒pSE1-3-9kb上,tet(X3.2)与tet(X3)和tet(X4)耐药表型类似[93]。He等[5]的流行病学研究显示,tet(X3)/tet(X4)在山东屠宰场的猪盲肠样本中检出率最高(40/60, 66.7%),替加环素耐药菌分离株主要为肠杆菌科(51/77)和不动杆菌属(16/77)。Dong等[94]分离得到289株tet(X)变异体阳性分离株,其中大部分不动杆菌分离株(83/289)携带tet(X3),所有大肠杆菌分离株(84/289)均携带tet(X4)。Sun等[20]从4 189份样本中鉴定出42株tet(X4)阳性的大肠杆菌分离株,其中有24株(24/42, 57.1%)大肠杆菌携带的tet(X4)位于质粒pLHM10-1-p6上,样本来源主要为猪粪便(30/4189, 0.7%),在污水、蔬菜和人身上均未检出。Li等[95]采用全基因组测序(WGS)和单质粒分子分析方法,研究了tet(X4)载体质粒的多样性,携带tet(X4)的质粒大小范围为9~294 kb,质粒种类有ColE2-like、IncQ、IncX1、IncA/C2、IncFII、IncFIB和具有不同复制子的杂交质粒。2020年,Fang等[2]的流行性病学回溯性研究表明,tet(X3)和tet(X4)已广泛分布于不动杆菌属和肠杆菌科,样本来源于动物、人、食物和环境中。
表2 tet(X)基因在不同来源细菌或环境中的流行情况
Table 2 Distribution of tet(X) genes in bacteria or environments from different sources
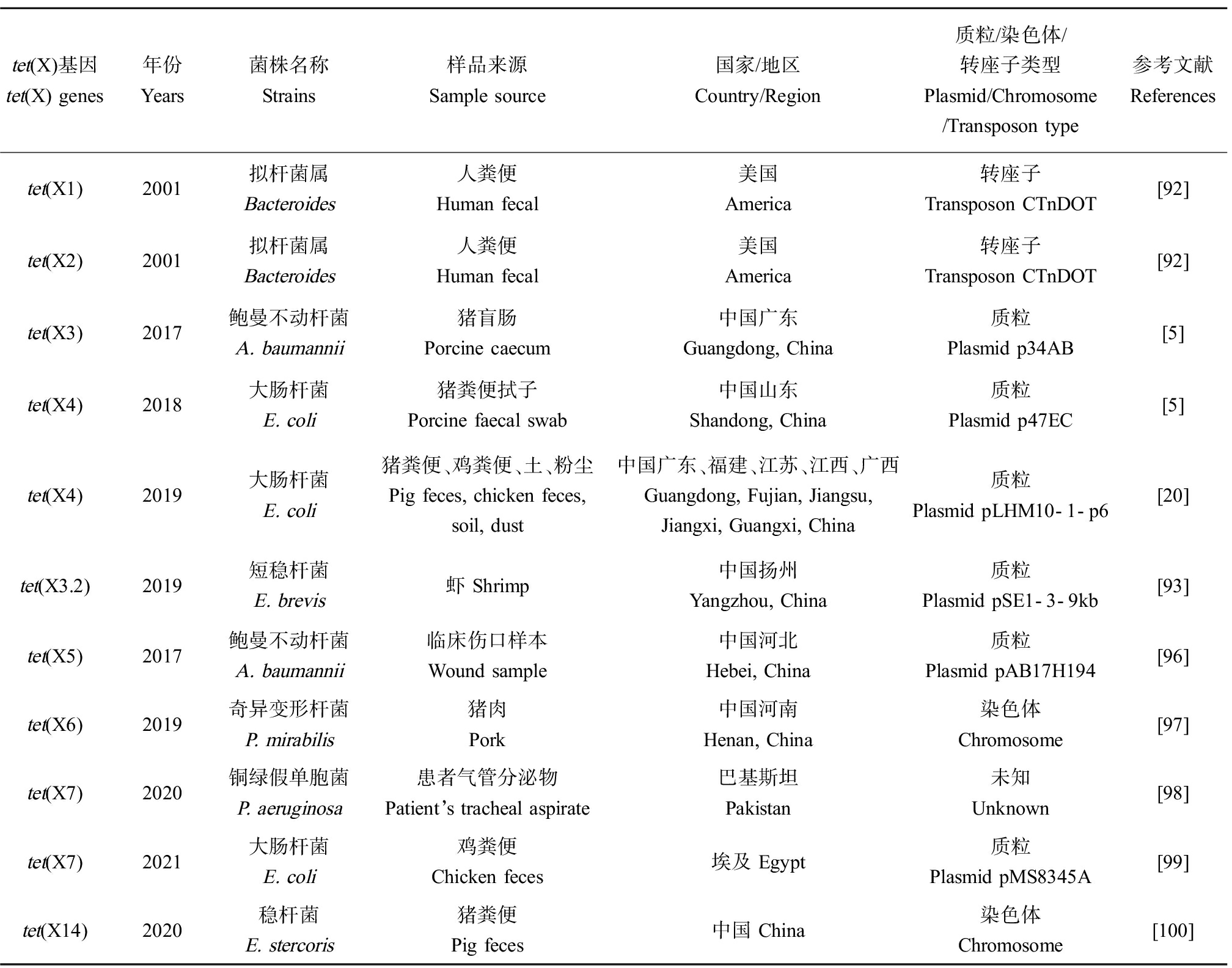
tet(X)基因tet(X) genes年份Years菌株名称Strains样品来源Sample source国家/地区Country/Region 质粒/染色体/转座子类型Plasmid/Chromosome /Transposon type参考文献Referencestet(X1)2001拟杆菌属 Bacteroides人粪便 Human fecal美国 America转座子Transposon CTnDOT[92]tet(X2)2001拟杆菌属 Bacteroides人粪便 Human fecal美国 America转座子Transposon CTnDOT[92]tet(X3)2017鲍曼不动杆菌 A. baumannii猪盲肠 Porcine caecum中国广东 Guangdong, China质粒 Plasmid p34AB[5]tet(X4)2018大肠杆菌 E. coli猪粪便拭子 Porcine faecal swab中国山东Shandong, China质粒 Plasmid p47EC[5]tet(X4)2019大肠杆菌 E. coli猪粪便、鸡粪便、土、粉尘Pig feces, chicken feces, soil, dust中国广东、福建、江苏、江西、广西Guangdong, Fujian, Jiangsu, Jiangxi, Guangxi, China质粒Plasmid pLHM10-1-p6[20]tet(X3.2)2019短稳杆菌 E. brevis虾 Shrimp中国扬州Yangzhou, China质粒 Plasmid pSE1-3-9kb[93]tet(X5)2017鲍曼不动杆菌 A. baumannii临床伤口样本Wound sample中国河北Hebei, China质粒 Plasmid pAB17H194[96]tet(X6)2019奇异变形杆菌 P. mirabilis猪肉 Pork中国河南Henan, China染色体Chromosome[97]tet(X7)2020铜绿假单胞菌P. aeruginosa患者气管分泌物Patient’s tracheal aspirate巴基斯坦Pakistan未知 Unknown[98]tet(X7)2021大肠杆菌 E. coli鸡粪便 Chicken feces埃及 Egypt质粒 Plasmid pMS8345A[99]tet(X14)2020稳杆菌E. stercoris猪粪便 Pig feces中国 China染色体 Chromosome[100]
2.2.4 tet(X5)~tet(X14)
tet(X5)首次发现于一株来源于临床伤口样本的鲍曼不动杆菌,tet(X5)位于质粒pAB17H194上,其耐药水平略低于tet(X3)和tet(X4)[96]。He等[97]从猪肉样本中分离出1株携带tet(X6)的奇异变形杆菌,tet(X6)位于染色体ICEPgs6Chn1,该染色体上还存在其他耐药基因floR、strB、strA、aph(30)-Ia、aac(3)-IV、aph(4)-Ia和sul2。Gasparrini等[98]从一株临床铜绿假单胞菌分离株中鉴定出tet(X7),该菌株来源于囊性纤维化患者气管吸出物。在同一项研究中,7个tet(X)变体tet(X7)~tet(X13)通过功能宏基因组学的方法检测出,样本来源于环境、人类共生菌和致病菌,这些基因介导包括FDA最近批准的奥马环素和依拉环素在内的所有四环素类抗生素的耐药性。2021年,Soliman等[99]从埃及鸡粪便样本中分离出5株携带tet(X7)的大肠杆菌菌株,该基因位于质粒pMS8345A上,药敏结果表明5株分离株对替加环素的MIC值为8~16 μg·mL-1。Cheng等[100]发现tet(X)家族新变体tet(X14),该基因位于稳杆菌的染色体上,菌株来源于中国养殖场的猪粪便样本,该项研究表明黄杆菌科可能是tet(X14)的主要宿主。
2.2.5 tet(X15)~tet(X17)
2020年,Liu等[101]在肉鸡和猪样本中发现了tet(X6)的4个亚型,亚型一来源于褐色类香味菌(Myroides phaeus),亚型二来源于卡氏变形杆菌(Proteus cibarius),亚型三来源于奇异变形杆菌(Proteus mirabilis),亚型四来源于鲁氏不动杆菌(Acinetobacter lwoffii)和约氏不动杆菌(Acinetobacter johnsonii),亚型二、三、四与亚型一的蛋白质序列差异分别为16、4和11个氨基酸。2021年,Xu等[102]将亚型二、三、四重新命名为tet(X15)~tet(X17)。
2.2.6 tet(X18)~tet(X44)
2021年,Umar等[103]从GenBank中收集的57个鸭疫里氏杆菌非冗余基因组序列中发现了26个新变异体tet(X18)~tet(X44),广泛扩展了现有tet(X)家族,研究结果表明鸭疫里氏杆菌是tet(X)基因家族的天然存储库。此外,该项研究还在鸭疫里氏杆菌的染色体中发现了Tet(X2)与Tet(X4)的进化前体Tet(X2/4)-P,对替加环素呈现中水平耐药(MIC=8 μg·mL-1)。鸭疫里氏杆菌是tet(X)基因家族传播到临床病原体的跳板[104]。
2.2.7 tet(X45)~tet(X47)
2021年,Zhang等[23]从12 829份人体微生物宏基因组样本中重建了202 265个宏基因组组装基因组(metagenome-assembled genomes, MAGs),并鉴定出3个新tet(X)基因,tet(X45)、tet(X46)和tet(X47)编码的蛋白与tet(X2)编码的蛋白分别有91%、97%和98%的氨基酸同源性,tet(X45)、tet(X46)和tet(X47)均对替加环素耐药(8~16 μg·mL-1),并对奥马环素(16~32 μg·mL-1)和依拉环素(2~4 μg·mL-1)高水平耐药。
综上所述,替加环素耐药机制已经得到国内外研究人员的持续关注[2, 104-106],目前已探明质粒介导的替加环素耐药机制有:RND型外排泵OqxAB和TMEXcd1-TOprJ1,MFS外排泵Tet(A)突变体和Tet(L)突变体,以及Tet(X)修饰酶及其变异体Tet(X3)、Tet(X4)、Tet(X5)、Tet(X6)和Tet(X7)。质粒介导的高水平替加环素耐药基因tet(X3)和tet(X4)被首次发现后,tet(X)变异体正以飞快的速度变化发展,目前已发展至tet(X47)。
2.3 tet(X)基因家族分布特征
为了解析tet(X)基因家族的细菌宿主以及质粒介导的替加环素耐药基因的分布情况,基于Zhang等[23]对tet(X)及其变异体的全球溯源分析,本研究从NCBI数据库中下载了852个携带替加环素耐药基因的细菌基因组加以分析。使用基因组分类数据库(Genome Taxonomy Database, GTDB)进行物种分类[107],使用耐药基因数据库(Comprehensive Antibiotic Resistance Database, CARD)注释细菌基因组上的耐药基因[108]。结果表明,tet(X)基因家族宿主主要为拟杆菌门(Bacteroidota)、弯曲杆菌门(Campylobacterota)、厚壁菌门(Firmicutes_A)和变形菌门(Proteobacteria)。在细菌科水平上分属于以下20种细菌:Acutalibacteraceae、Aeromonadaceae、Arcobacteraceae、Bacteroidaceae、Campylobacteraceae、Chitinophagaceae、Enterobacteriaceae、Flavobacteriaceae、Lachnospiraceae、Marinifilaceae、Moraxellaceae、Muribaculaceae、Paludibacteraceae、Pseudomonadaceae、Rikenellaceae、Saprospiraceae、Shewanellaceae、Sphingobacteriaceae、Tannerellaceae和Weeksellaceae。其中,肠杆菌科(Enterobacteriaceae)、莫拉氏菌科不动杆菌属(Moraxellaceae,Acinetobacter)、拟杆菌科(Bacteroidaceae)、黄杆菌科(Flavobacteriaceae)、威克氏菌科(Weeksellaceae)多种细菌携带替加环素耐药基因(图4(a))。在细菌基因组水平上,852株细菌的染色体和质粒上携带tet(X)、tet(X1)、tet(X3)、tet(X4)、tet(X5)和tet(X6),同时共存多种四环素类抗生素耐药基因:tet(33)、tet(39)、tet(40)、tet(43)、tet(45)、tet(49)、tet(52)、tet(A)、tet(B)、tet(C)、tet(D)、tet(J)、tet(V)、tet(W/N/W)、tet(X1)、tet(X3)、tet(X4)、tet(X5)、tet(X6)、tet(Y)、tet32、tet36、tetA(P)、tetB(60)和tetB(P)(图4(b))。此外,852个细菌基因组中共有74个质粒携带四环素耐药基因,这些质粒来自不动杆菌属10种不同细菌,如肠杆菌科大肠杆菌、费格森埃希菌、变形杆菌,拟杆菌细菌等,质粒上总共发现13种四环素耐药基因:tet(A)、tet(B)、tet(D)、tet(X3)、tet(X4)、tet(X5)、tet(X6)、tet(Y)、tet36、tetB(60)、tetM、tetR和tet(X)(图4(b))。值得注意的是,这些菌株同时携带不同类型的临床重要抗生素耐药基因,如超广谱β-内酰胺酶基因(blaCMY、blaCTX-M、blaIMP、blaKPC、blaNDM、blaOXA、blaSHV和blaTEM)、质粒介导的粘菌素耐药基因(mcr-1.1、mcr-3.5、mcr-1.4、mcr-3.2、mcr-3.1和mcr-9.1)。

图4 tet(X)宿主细菌物种分类及其携带的耐药基因
注:(a) tet(X)宿主细菌物种分类;(b) tet(X)在细菌染色体和质粒中的分布特征。
Fig. 4 Phylogenetic analyses of the tet(X) carrying isolates
Note: (a) Genome taxonomy of tet(X) carrying isolates; (b) Prevalence of tet(X) in bacterial genome and plasmids.
在852个携带替加环素耐药基因的细菌中,一株携带tet(X)基因的超级耐药致病菌值得关注。Hu等[109]从病人尿液样本中分离出一株多耐药香味类香味菌(Myroides odoratimimus)PR63039,抗生素敏感性试验结果显示PR63039对所测试的21种抗生素均耐药。基因组分析显示其基因组含有大量的耐药基因,包括β-内酰胺酶、四环素、氯霉素、喹诺酮、多耐药质子泵基因、耐药突变基因rpoB和gyrA,并且染色体和质粒上均携带有tet(X)基因。从病人身上分离出来的携带tet(X)基因的超级耐药菌的出现,对抗生素疗效构成极大的威胁,应警惕此类携带tet(X)及其变异体的超级耐药致病菌介导的替加环素高水平耐药性。
3 研究展望(Research prospect)
细菌耐药性问题已经对生态安全与公共健康构成重大威胁,世界各国正积极采取行动,从动物-环境-人类的视角,以“大健康(One Health)”理念开展遏制细菌耐药工作。细菌基因组数据已证明替加环素耐药基因的细菌宿主同时携带不同类型的临床重要抗生素耐药基因,表明其耐药机制趋于复杂。因此,研究替加环素的耐药机制和传播机制,对临床治疗多重耐药菌的感染和遏制细菌耐药性具有重要意义。对于今后的研究重点,我们认为需要主要关注几个方面的工作。
(1)超级耐药菌的出现是多种抗生素选择压力下共同作用的结果。虽然目前替加环素未被批准用于养殖行业,但替加环素耐药可导致第四代新型四环素类药物的交叉耐药,同时影响一代、二代四环素类抗生素的应用,仍应持续关注养殖业中新型替加环素耐药机制的产生与发展。
(2)替加环素高水平耐药基因的宿主属于不同细菌物种,并且分布于临床环境和非临床环境,可能会在“动物-环境-人类”链条中传播,当前耐药细菌的传播机制研究主要集中于临床环境,应加强非临床环境(如养殖环境和城市环境)中的研究。
(3)2020年后tet(X)基因家族的不断壮大归因于宏基因组学技术的发展,tet(X)变异体成员的迅速增加说明了其大规模传播的可能性,因此,有必要进一步通过宏基因组学监测新发现的tet(X)变异体的快速传播,并探究其他未知的四环素修饰酶基因,重点监测新的四环素特异性外排泵突变体介导的替加环素耐药性。
[1] McAllister T A, Wang Y X, Diarra M S, et al. Challenges of a one-health approach to the development of alternatives to antibiotics [J]. Animal Frontiers: The Review Magazine of Animal Agriculture, 2018, 8(2): 10-20
[2] Fang L X, Chen C, Cui C Y, et al. Emerging high-level tigecycline resistance: Novel tetracycline destructases spread via the mobile Tet(X) [J]. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 2020, 42(8): e2000014
[3] Michalova E, Novotna P, Schlegelova J. Tetracyclines in veterinary medicine and bacterial resistance to them [J]. Veterinarni Medicina, 2004, 49(3): 79-100
[4] Daghrir R, Drogui P. Tetracycline antibiotics in the environment: A review [J]. Environmental Chemistry Letters, 2013, 11(3): 209-227
[5] He T, Wang R, Liu D J, et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans [J]. Nature Microbiology, 2019, 4(9): 1450-1456
[6] Roberts M C, Schwarz S. Tetracycline and phenicol resistance genes and mechanisms: Importance for agriculture, the environment, and humans [J]. Journal of Environmental Quality, 2016, 45(2): 576-592
[7] Izaki K, Arima K. Disappearance of oxytetracycline accumulation in the cells of multiple drug-resistant Escherichia coli [J]. Nature, 1963, 200: 384-385
[8] Levy S B, McMurry L. Plasmid-determined tetracycline resistance involves new transport systems for tetracycline [J]. Nature, 1978, 276(5683): 90-92
[9] Kaneko M, Yamaguchi A, Sawai T. Energetics of tetracycline efflux system encoded by Tn10 in Escherichia coli [J]. FEBS Letters, 1985, 193(2): 194-198
[10] Mendez B, Tachibana C, Levy S B. Heterogeneity of tetracycline resistance determinants [J]. Plasmid, 1980, 3(2): 99-108
[11] Burdett V. Streptococcal tetracycline resistance mediated at the level of protein synthesis [J]. Journal of Bacteriology, 1986, 165(2): 564-569
[12] Burdett V. Purification and characterization of Tet(M), a protein that renders ribosomes resistant to tetracycline [J]. The Journal of Biological Chemistry, 1991, 266(5): 2872-2877
[13] Burdett V. Tet(M)-promoted release of tetracycline from ribosomes is GTP dependent [J]. Journal of Bacteriology, 1996, 178(11): 3246-3251
[14] Yang W R, Moore I F, Koteva K P, et al. TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics [J]. Journal of Biological Chemistry, 2004, 279(50): 52346-52352
[15] Grossman T H. Tetracycline antibiotics and resistance [J]. Cold Spring Harbor Perspectives in Medicine, 2016, 6(4): a025387
[16] Zhanel G G, Homenuik K, Nichol K, et al. The glycylcyclines: A comparative review with the tetracyclines [J]. Drugs, 2004, 64(1): 63-88
[17] Yaghoubi S, Zekiy A O, Krutova M, et al. Tigecycline antibacterial activity, clinical effectiveness, and mechanisms and epidemiology of resistance: Narrative review [J]. European Journal of Clinical Microbiology & Infectious Diseases, 2022, 41(7): 1003-1022
[18] World Health Organization. Critically important antimicrobials for human medicine 2019 [R]. Geneva: World Health Organization, 2019
[19] Chen C, Cui C Y, Yu J J, et al. Genetic diversity and characteristics of high-level tigecycline resistance Tet(X) in Acinetobacter species [J]. Genome Medicine, 2020, 12(1): 111
[20] Sun J, Chen C, Cui C Y, et al. Plasmid-encoded Tet(X) genes that confer high-level tigecycline resistance in Escherichia coli [J]. Nature Microbiology, 2019, 4(9): 1457-1464
[21] Sun C T, Cui M Q, Zhang S, et al. Plasmid-mediated tigecycline-resistant gene Tet(X4) in Escherichia coli from food-producing animals, China, 2008-2018 [J]. Emerging Microbes & Infections, 2019, 8(1): 1524-1527
[22] Sun C T, Cui M Q, Zhang S, et al. Genomic epidemiology of animal-derived tigecycline-resistant Escherichia coli across China reveals recent endemic plasmid-encoded Tet(X4) gene [J]. Communications Biology, 2020, 3: 412
[23] Zhang R M, Sun J, Sun R Y, et al. Source tracking and global distribution of the tigecycline non-susceptible tet(X) [J]. Microbiology Spectrum, 2021, 9(3): e0116421
[24] Livermore D M. Tigecycline: What is it, and where should it be used? [J]. The Journal of Antimicrobial Chemotherapy, 2005, 56(4): 611-614
[25] Bradford P A. Tigecycline: A first in class glycylcycline [J]. Clinical Microbiology Newsletter, 2004, 26(21): 163-168
[26] Dougherty J A, Sucher A J, Chahine E B, et al. Omadacycline: A new tetracycline antibiotic [J]. The Annals of Pharmacotherapy, 2019, 53(5): 486-500
[27] Zhanel G G, Cheung D, Adam H, et al. Review of eravacycline, a novel fluorocycline antibacterial agent [J]. Drugs, 2016, 76(5): 567-588
[28] 孙广龙, 胡立宏. 四环素类抗生素的研究进展[J]. 药学研究, 2017, 36(1): 1-5
Sun G L, Hu L H. Advances in the research of tetracyclines antibiotics [J]. Journal of Pharmaceutical Research, 2017, 36(1): 1-5 (in Chinese)
[29] Verchère A, Broutin I, Picard M. Photo-induced proton gradients for the in vitro investigation of bacterial efflux pumps [J]. Scientific Reports, 2012, 2: 306
[30] Adam Y, Tayer N, Rotem D, et al. The fast release of sticky protons: Kinetics of substrate binding and proton release in a multidrug transporter [J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(46): 17989-17994
[31] Zechini B, Versace I. Inhibitors of multidrug resistant efflux systems in bacteria [J]. Recent Patents on Anti-Infective Drug Discovery, 2009, 4(1): 37-50
[32] 李晶, 付喜爱, 刘耀川, 等. 细菌四环素类药物外排泵的研究新进展[J]. 黑龙江畜牧兽医, 2014(23): 48-50
Li J, Fu X A, Liu Y C, et al. New research advances in tetracycline drug efflux pump in bacteria [J]. Heilongjiang Animal Science and Veterinary Medicine, 2014(23): 48-50 (in Chinese)
[33] Poole K. Efflux-mediated antimicrobial resistance [J]. The Journal of Antimicrobial Chemotherapy, 2005, 56(1): 20-51
[34] Thaker M, Spanogiannopoulos P, Wright G D. The tetracycline resistome [J]. Cellular and Molecular Life Sciences, 2010, 67(3): 419-431
[35] Forsberg K J, Patel S, Wencewicz T A, et al. The tetracycline destructases: A novel family of tetracycline-inactivating enzymes [J]. Chemistry & Biology, 2015, 22(7): 888-897
[36] McGivern B B, McDonell R K, Morris S K, et al. Novel class 1 integron harboring antibiotic resistance genes in wastewater-derived bacteria as revealed by functional metagenomics [J]. Plasmid, 2021, 114: 102563
[37] Zhu Y, Wang C Z, Schwarz S, et al. Identification of a novel tetracycline resistance gene, Tet(63), located on a multiresistance plasmid from Staphylococcus aureus [J]. The Journal of Antimicrobial Chemotherapy, 2021, 76(3): 576-581
[38] Somprasong N, Hall C M, Webb J R, et al. Burkholderia ubonensis high-level tetracycline resistance is due to efflux pump synergy involving a novel TetA(64) resistance determinant [J]. Antimicrobial Agents and Chemotherapy, 2021, 65(3): e01767-e01720
[39] Bergeron J, Ammirati M, Danley D, et al. Glycylcyclines bind to the high-affinity tetracycline ribosomal binding site and evade Tet(M)- and Tet(O)-mediated ribosomal protection [J]. Antimicrobial Agents and Chemotherapy, 1996, 40(9): 2226-2228
[40] Stein G E, Babinchak T. Tigecycline: An update [J]. Diagnostic Microbiology and Infectious Disease, 2013, 75(4): 331-336
[41] Hoban D J, Bouchillon S K, Johnson B M, et al. In vitro activity of tigecycline against 6792 Gram-negative and Gram-positive clinical isolates from the global Tigecycline Evaluation and Surveillance Trial (TEST Program, 2004) [J]. Diagnostic Microbiology and Infectious Disease, 2005, 52(3): 215-227
[42] Pournaras S, Koumaki V, Spanakis N, et al. Current perspectives on tigecycline resistance in Enterobacteriaceae: Susceptibility testing issues and mechanisms of resistance [J]. International Journal of Antimicrobial Agents, 2016, 48(1): 11-18
[43] Jiang Y S, Yang S S, Deng S L, et al. Epidemiology and resistance mechanisms of tigecycline- and carbapenem-resistant Enterobacter cloacae in Southwest China: A 5-year retrospective study [J]. Journal of Global Antimicrobial Resistance, 2022, 28: 161-167
[44] Sun Y, Cai Y, Liu X, et al. The emergence of clinical resistance to tigecycline [J]. International Journal of Antimicrobial Agents, 2013, 41(2): 110-116
[45] Villa L, Feudi C, Fortini D, et al. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance [J]. Antimicrobial Agents and Chemotherapy, 2014, 58(3): 1707-1712
[46] Beabout K, Hammerstrom T G, Perez A M, et al. The ribosomal S10 protein is a general target for decreased tigecycline susceptibility [J]. Antimicrobial Agents and Chemotherapy, 2015, 59(9): 5561-5566
[47] Cattoir V, Isnard C, Cosquer T, et al. Genomic analysis of reduced susceptibility to tigecycline in Enterococcus faecium [J]. Antimicrobial Agents and Chemotherapy, 2015, 59(1): 239-244
[48] Li X, Liu L, Ji J, et al. Tigecycline resistance in Acinetobacter baumannii mediated by frameshift mutation in plsC, encoding 1-acyl-sn-glycerol-3-phosphate acyltransferase [J]. European Journal of Clinical Microbiology & Infectious Diseases, 2015, 34(3): 625-631
[49] Li X, Quan J, Yang Y, et al. Abrp, a new gene, confers reduced susceptibility to tetracycline, glycylcine, chloramphenicol and fosfomycin classes in Acinetobacter baumannii [J]. European Journal of Clinical Microbiology & Infectious Diseases: Official Publication of the European Society of Clinical Microbiology, 2016, 35(8): 1371-1375
[50] Yu R, Zhang Y, Xu Y D, et al. Emergence of a tet(M) variant conferring resistance to tigecycline in Streptococcus suis [J]. Frontiers in Veterinary Science, 2021, 8: 709327
[51] Webber M A, Piddock L J. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli [J]. Antimicrobial Agents and Chemotherapy, 2001, 45(5): 1550-1552
[52] Hornsey M, Ellington M J, Doumith M, et al. Tigecycline resistance in Serratia marcescens associated with up-regulation of the SdeXY-HasF efflux system also active against ciprofloxacin and cefpirome [J]. The Journal of Antimicrobial Chemotherapy, 2010, 65(3): 479-482
[53] Veleba M, De Majumdar S, Hornsey M, et al. Genetic characterization of tigecycline resistance in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes [J]. The Journal of Antimicrobial Chemotherapy, 2013, 68(5): 1011-1018
[54] Lv L C, Wan M, Wang C Z, et al. Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in Klebsiella pneumoniae [J]. mBio, 2020, 11(2): e02930-e02919
[55] Pournaras S, Koumaki V, Gennimata V, et al. In vitro activity of tigecycline against Acinetobacter baumannii: Global epidemiology and resistance mechanisms [J]. Advances in Experimental Medicine and Biology, 2016, 897: 1-14
[56] Keeney D, Ruzin A, McAleese F, et al. MarA-mediated overexpression of the AcrAB efflux pump results in decreased susceptibility to tigecycline in Escherichia coli [J]. The Journal of Antimicrobial Chemotherapy, 2008, 61(1): 46-53
[57] He F, Fu Y, Chen Q, et al. Tigecycline susceptibility and the role of efflux pumps in tigecycline resistance in KPC-producing Klebsiella pneumoniae [J]. PLoS One, 2015, 10(3): e0119064
[58] Veleba M, Higgins P G, Gonzalez G, et al. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae [J]. Antimicrobial Agents and Chemotherapy, 2012, 56(8): 4450-4458
[59] Jiménez-Castellanos J C, Wan Ahmad Kamil W N, Cheung C H P, et al. Comparative effects of overproducing the AraC-type transcriptional regulators MarA, SoxS, RarA and RamA on antimicrobial drug susceptibility in Klebsiella pneumoniae [J]. The Journal of Antimicrobial Chemotherapy, 2016, 71(7): 1820-1825
[60] Chen J, Kuroda T, Huda M N, et al. An RND-type multidrug efflux pump SdeXY from Serratia marcescens [J]. The Journal of Antimicrobial Chemotherapy, 2003, 52(2): 176-179
[61] Kumar A, Worobec E A. HasF, a TolC-homolog of Serratia marcescens, is involved in energy-dependent efflux [J]. Canadian Journal of Microbiology, 2005, 51(6): 497-500
[62] Li R C, Peng K, Xiao X, et al. Emergence of a multidrug resistance efflux pump with carbapenem resistance gene blaVIM-2 in a Pseudomonas putida megaplasmid of migratory bird origin [J]. The Journal of Antimicrobial Chemotherapy, 2021, 76(6): 1455-1458
[63] He R W, Yang Y Q, Wu Y P, et al. Characterization of a plasmid-encoded resistance-nodulation-division efflux pump in Klebsiella pneumoniae and Klebsiella quasipneumoniae from patients in China [J]. Antimicrobial Agents and Chemotherapy, 2021, 65(2): e02075-e02020
[64] Hirabayashi A, Ha V, Nguyen A V, et al. Emergence of a plasmid-borne tigecycline resistance in Klebsiella pneumoniae in Vietnam [J]. Journal of Medical Microbiology, 2021, 70(3): 10.1099/jmm.0.001320
[65] Wan M, Gao X, Lv L C, et al. IS 26 mediates the acquisition of tigecycline resistance gene cluster tmexCD1-toprJ1 by IncHI1B-FIB plasmids in Klebsiella pneumoniae and Klebsiella quasipneumoniae from food market sewage [J]. Antimicrobial Agents and Chemotherapy, 2021, 65(3): e02178-e02120
[66] Wang C Z, Gao X, Yang Q W, et al. A novel transferable resistance-nodulation-division pump gene cluster, tmexCD2-toprJ2, confers tigecycline resistance in Raoultella ornithinolytica [J]. Antimicrobial Agents and Chemotherapy, 2021, 65(4): e02229-e02220
[67] Wang Q, Peng K, Liu Y, et al. Characterization of TMexCD3-TOprJ3, an RND-type efflux system conferring resistance to tigecycline in Proteus mirabilis, and its associated integrative conjugative element [J]. Antimicrobial Agents and Chemotherapy, 2021, 65(7): e0271220
[68] Hirabayashi A, Dao T D, Takemura T, et al. A transferable IncC-IncX3 hybrid plasmid cocarrying blaNDM-4, tet(X), and tmexCD3-toprJ3 confers resistance to carbapenem and tigecycline [J]. mSphere, 2021, 6(4): e0059221
[69] Peleg A Y, Adams J, Paterson D L. Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii [J]. Antimicrobial Agents and Chemotherapy, 2007, 51(6): 2065-2069
[70] Lucaßen K, Müller C, Wille J, et al. Prevalence of RND efflux pump regulator variants associated with tigecycline resistance in carbapenem-resistant Acinetobacter baumannii from a worldwide survey [J]. The Journal of Antimicrobial Chemotherapy, 2021, 76(7): 1724-1730
[71] Yang Y S, Chen H Y, Hsu W J, et al. Overexpression of AdeABC efflux pump associated with tigecycline resistance in clinical Acinetobacter nosocomialis isolates [J]. Clinical Microbiology and Infection, 2019, 25(4): 512.e1-512.e6
[72] Sengeløv G, Halling-Sørensen B, Aarestrup F M. Susceptibility of Escherichia coli and Enterococcus faecium isolated from pigs and broiler chickens to tetracycline degradation products and distribution of tetracycline resistance determinants in E. coli from food animals [J]. Veterinary Microbiology, 2003, 95(1-2): 91-101
[73] Møller T S B, Overgaard M, Nielsen S S, et al. Relation between tetR and tetA expression in tetracycline resistant Escherichia coli [J]. BMC Microbiology, 2016, 16: 39
[74] Hentschke M, Christner M, Sobottka I, et al. Combined ramR mutation and presence of a Tn1721-associated Tet(A) variant in a clinical isolate of Salmonella enterica serovar Hadar resistant to tigecycline [J]. Antimicrobial Agents and Chemotherapy, 2010, 54(3): 1319-1322
[75] Xu J, Zhu Z L, Chen Y M, et al. The plasmid-borne tet(A) gene is an important factor causing tigecycline resistance in ST11 carbapenem-resistant Klebsiella pneumoniae under selective pressure [J]. Frontiers in Microbiology, 2021, 12: 644949
[76] Chiu S K, Huang L Y, Chen H, et al. Roles of ramR and tet(A) mutations in conferring tigecycline resistance in carbapenem-resistant Klebsiella pneumoniae clinical isolates [J]. Antimicrobial Agents and Chemotherapy, 2017, 61(8): e00391-e00317
[77] De Jesus M, Jin J, Guffanti A A, et al. Importance of the GP dipeptide of the antiporter motif and other membrane-embedded proline and glycine residues in tetracycline efflux protein Tet(L) [J]. Biochemistry, 2005, 44(38): 12896-12904
[78] Ginn S L, Brown M H, Skurray R A. Membrane topology of the metal-tetracycline/H+ antiporter TetA(K) from Staphylococcus aureus [J]. Journal of Bacteriology, 1997, 179(11): 3786-3789
[79] Jin J, Guffanti A A, Bechhofer D H, et al. Tet(L) and Tet(K) tetracycline-divalent metal/H+ antiporters: Characterization of multiple catalytic modes and a mutagenesis approach to differences in their efflux substrate and coupling ion preferences [J]. Journal of Bacteriology, 2002, 184(17): 4722-4732
[80] Schwarz S, Cardoso M, Wegener H C. Nucleotide sequence and phylogeny of the Tet(L) tetracycline resistance determinant encoded by plasmid pSTE1 from Staphylococcus hyicus [J]. Antimicrobial Agents and Chemotherapy, 1992, 36(3): 580-588
[81] Jiang N S, Li J, Feßler A T, et al. A novel small Tet(T)-Tet(L)-aadD-carrying plasmid from MRSA and MSSA ST9 isolates of swine origin [J]. The Journal of Antimicrobial Chemotherapy, 2019, 74(8): 2462-2464
[82] Chopra I, Roberts M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance [J]. Microbiology and Molecular Biology Reviews, 2001, 65(2): 232-260
[83] Kehrenberg C, Catry B, Haesebrouck F, et al. Tet(L)-mediated tetracycline resistance in bovine Mannheimia and Pasteurella isolates [J]. The Journal of Antimicrobial Chemotherapy, 2005, 56(2): 403-406
[84] Fiedler S, Bender J K, Klare I, et al. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants Tet(L) and Tet(M) [J]. The Journal of Antimicrobial Chemotherapy, 2016, 71(4): 871-881
[85] Wang N N, Li D X, Schwarz S, et al. Novel Tet(L) efflux pump variants conferring resistance to tigecycline and eravacycline in Staphylococcus spp. [J]. Microbiology Spectrum, 2021, 9(3): e0131021
[86] Yao H, Jiao D, Zhao W B, et al. Emergence of a novel tet(L) variant in Campylobacter spp. of chicken origin in China [J]. Antimicrobial Agents and Chemotherapy, 2020, 65(1): e01622-e01620
[87] Tally F P, Snydman D R, Shimell M J, et al. Characterization of pBFTM10, a clindamycin-erythromycin resistance transfer factor from Bacteroides fragilis [J]. Journal of Bacteriology, 1982, 151(2): 686-691
[88] Guiney D G Jr, Hasegawa P, Davis C E. Expression in Escherichia coli of cryptic tetracycline resistance genes from Bacteroides R plasmids [J]. Plasmid, 1984, 11(3): 248-252
[89] Matthews B G, Guiney D G. Characterization and mapping of regions encoding clindamycin resistance, tetracycline resistance, and a replication function on the Bacteroides R plasmid pCP1 [J]. Journal of Bacteriology, 1986, 167(2): 517-521
[90] Speer B S, Salyers A A. Novel aerobic tetracycline resistance gene that chemically modifies tetracycline [J]. Journal of Bacteriology, 1989, 171(1): 148-153
[91] Speer B S, Salyers A A. A tetracycline efflux gene on Bacteroides transposon Tn4400 does not contribute to tetracycline resistance [J]. Journal of Bacteriology, 1990, 172(1): 292-298
[92] Whittle G, Hund B D, Shoemaker N B, et al. Characterization of the 13-kilobase ermF region of the Bacteroides conjugative transposon CTnDOT [J]. Applied and Environmental Microbiology, 2001, 67(8): 3488-3495
[93] Li R C, Liu Z Y, Peng K, et al. Co-occurrence of two tet(X) variants in an Empedobacter brevis of shrimp origin [J]. Antimicrobial Agents and Chemotherapy, 2019, 63(12): e01636-e01619
[94] Dong N, Zeng Y, Cai C, et al. Prevalence, transmission, and molecular epidemiology of Tet(X)-positive bacteria among humans, animals, and environmental niches in China: An epidemiological, and genomic-based study [J]. The Science of the Total Environment, 2022, 818: 151767
[95] Li R C, Lu X Y, Peng K, et al. Deciphering the structural diversity and classification of the mobile tigecycline resistance gene tet(X)-bearing plasmidome among bacteria [J]. mSystems, 2020, 5(2): e00134-e00120
[96] Wang L Y, Liu D J, Lv Y, et al. Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline, and omadacycline in a clinical Acinetobacter baumannii isolate [J]. Antimicrobial Agents and Chemotherapy, 2019, 64(1): e01326-e01319
[97] He D D, Wang L L, Zhao S Y, et al. A novel tigecycline resistance gene, Tet(X6), on an SXT/R391 integrative and conjugative element in a Proteus genomospecies 6 isolate of retail meat origin [J]. The Journal of Antimicrobial Chemotherapy, 2020, 75(5): 1159-1164
[98] Gasparrini A J, Markley J L, Kumar H, et al. Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance [J]. Communications Biology, 2020, 3: 241
[99] Soliman A M, Ramadan H, Zarad H, et al. Coproduction of Tet(X7) conferring high-level tigecycline resistance, fosfomycin FosA4, and colistin mcr-1.1 in Escherichia coli strains from chickens in Egypt [J]. Antimicrobial Agents and Chemotherapy, 2021, 65(6): e02084-e02020
[100] Cheng Y Y, Chen Y, Liu Y, et al. Identification of novel tetracycline resistance gene Tet(X14) and its co-occurrence with Tet(X2) in a tigecycline-resistant and colistin-resistant Empedobacter stercoris [J]. Emerging Microbes & Infections, 2020, 9(1): 1843-1852
[101] Liu D J, Zhai W S, Song H W, et al. Identification of the novel tigecycline resistance gene Tet(X6) and its variants in Myroides, Acinetobacter and Proteus of food animal origin [J]. The Journal of Antimicrobial Chemotherapy, 2020, 75(6): 1428-1431
[102] Xu Y C, Liu L Z, Zhang H M, et al. Co-production of Tet(X) and MCR-1, two resistance enzymes by a single plasmid [J]. Environmental Microbiology, 2021, 23(12): 7445-7464
[103] Umar Z, Chen Q W, Tang B, et al. The poultry pathogen Riemerella anatipestifer appears as a reservoir for Tet(X) tigecycline resistance [J]. Environmental Microbiology, 2021, 23(12): 7465-7482
[104] Cui C Y, Chen Q W, He Q, et al. Transferability of tigecycline resistance: Characterization of the expanding Tet(X) family [J]. WIREs Mechanisms of Disease, 2022, 14(1): e1538
[105] 王知任, 李荷楠. 质粒介导的替加环素耐药机制的研究进展[J]. 中国感染与化疗杂志, 2021, 21(2): 230-234
Wang Z R, Li H N. Recent advances in plasmid-mediated mechanism of tigecycline resistance [J]. Chinese Journal of Infection and Chemotherapy, 2021, 21(2): 230-234 (in Chinese)
[106] 王亮亮, 王倩倩, 周博伦, 等. 替加环素耐药机制研究进展[J]. 中国畜牧兽医, 2020, 47(9): 3022-3029
Wang L L, Wang Q Q, Zhou B L, et al. Advance on the tigecycline resistance mechanism [J]. China Animal Husbandry & Veterinary Medicine, 2020, 47(9): 3022-3029 (in Chinese)
[107] Parks D H, Chuvochina M, Waite D W, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life [J]. Nature Biotechnology, 2018, 36(10): 996-1004
[108] Alcock B P, Raphenya A R, Lau T T Y, et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database [J]. Nucleic Acids Research, 2020, 48(D1): D517-D525
[109] Hu S H, Jiang T, Zhou Y J, et al. Genomic analysis of the multi-drug-resistant clinical isolate Myroides odoratimimus PR63039 [J]. Molecular Genetics and Genomics, 2017, 292(1): 133-144