随着规模化畜禽养殖的不断发展,畜禽粪便的处置成为重大问题[1]。其中,粪便作为肥料施加到土壤中是常见的处置方式[2]。施加粪肥可以增加土壤有机质及氮磷养分含量,提高作物产量[3-4]。但是畜禽粪便携带的丰富人类致病菌(human pathogen bacteria, HPB)也会污染土壤,通过直接接触或间接食物链传播危害人类健康安全[5-7]。同时,由于抗生素在畜禽养殖业的广泛使用,其诱导产生的抗生素抗性基因(antibiotic resistance genes, ARGs)可以在病原体间转移从而提高致病菌的环境适应能力[8]。研究发现粪肥处理土壤中抗性细菌得到富集[9]。Forsberg等[10]利用宏基因组测序方法,发现土壤中对多种抗生素具有耐药性的多重耐药细菌与多种病原菌具有相同的核酸序列,揭露了病原菌从土壤微生物中获取抗生素耐药性的可能。目前,对施用粪肥土壤中人类病原体传播的系统研究比较少。土壤中的病原菌可通过食物链或劳作时接触的方式进入人体。如果人类致病菌具有抗生素耐药性,发生细菌感染性疾病时药物将不起作用。因此,研究施加粪肥对土壤中人类致病菌变化的影响刻不容缓。
四川省畜禽养殖业发达,其粪便产生量位居全国前列[1]。紫色土是四川盆地重要的耕地土壤资源,其有机质含量低(<1.5%),结构差,常施以粪肥以改善土质。目前,关于施加粪肥后紫色土中病原菌变化的研究较少。因此,本研究以长期施用化肥紫色土作为实验对象,利用高通量测序技术,研究施加猪粪或鸡粪对土壤中人类致病菌的影响。主要回答3个问题:(1)施用粪肥土壤和未施肥土壤中人类致病菌的分布;(2)致病菌与ARGs的关系;(3)影响培养期间致病菌变化的因素。对施加粪肥后土壤中人类致病菌进行追踪调查,可为阻控人类致病菌传播提供一定的理论基础和有效科学依据。
1 材料与方法(Materials and methods)
1.1 温室盆钵实验
实验用土采自中国科学院盐亭紫色土农业生态试验站(31°16′ N,105°27′ E)内一处长期施用化肥样地。土壤风干,研磨,过2 mm筛后混合均匀。所用猪粪(pig manure, PM)、鸡粪(chicken manure, CM)取自当地养殖场,预处理方法与土壤相同。
实验共设有3个处理:未施肥处理(CK)、施加猪粪处理(PMs)和施加鸡粪处理(CMs),每种处理3个重复。粪肥施用量为土壤干质量的2%,各处理的每个花盆中土壤总质量均为2.5 kg。培养实验在温室中进行,培养过程中通过定期补充流失的水分使土壤含水率保持在70%田间持水量。
1.2 样品采集与理化分析
在第0、23、45、62和100天用土钻在花盆中进行多点取样,将每种处理的3个重复样混合成一个代表样,每种处理大约取15 g土壤样品,经冷冻干燥后过2 mm筛,-20 ℃保存。采用重铬酸钾氧化法和凯氏定氮法分别测定土壤有机碳(soil organic carbon, SOC)和总氮(total nitrogen, TN)的含量,结果如表1所示。
表1 不同采样时间点各处理的理化性质
Table 1 Physical and chemical properties of each treatment at different sampling time points
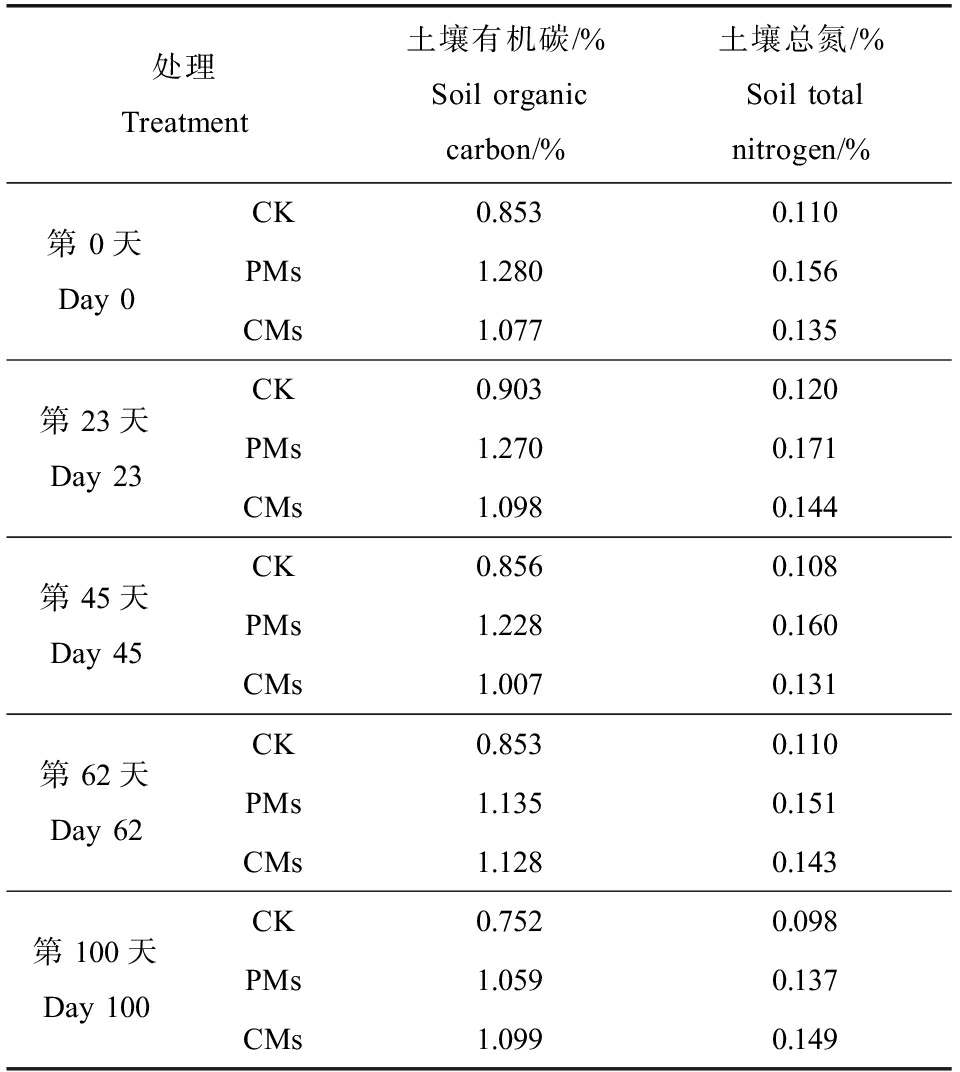
处理Treatment土壤有机碳/%Soil organic carbon/%土壤总氮/%Soil total nitrogen/%第0天Day 0CK0.8530.110PMs1.2800.156CMs1.0770.135第23天Day 23CK0.9030.120PMs1.2700.171CMs1.0980.144第45天Day 45CK0.8560.108PMs1.2280.160CMs1.0070.131第62天Day 62CK0.8530.110PMs1.1350.151CMs1.1280.143第100天Day 100CK0.7520.098PMs1.0590.137CMs1.0990.149
注:CK表示未施肥土壤;PMs表示施加猪粪土壤;CMs表示施加鸡粪土壤。
Note: CK means unfertilized soil; PMs means soil with pig manure; and CMs means soil with chicken manure.
1.3 土壤DNA提取
使用MoBio PowerSoil DNA提取试剂盒(MoBio Inc,Carlsbad,美国),按照说明书的步骤,从0.25 g土壤子样品中提取基因组DNA。得到的DNA样品用1%琼脂糖凝胶电泳检测完整性,并用微量核酸蛋白质分析仪(NanoDropTM One,Thermo Scientific,美国)检测DNA的纯度和浓度。A260/A280比值>1.8的DNA样品保存在-20 ℃冰箱以备后续扩增。
1.4 第二代测序和人类致病菌分析
用针对V4-V5区域的引物515F和909R扩增细菌的16S rRNA基因[11]。反向引物用独特的条形码标记以区分样品。PCR的步骤如下:94 ℃预变性3 min;30个热循环(94 ℃变性30 s,56 ℃退火60 s,72 ℃延伸60 s);最终在72 ℃延伸10 min。PCR扩增产物纯化后,采用Illumina Miseq平台进行测序。使用QIIME 1.9.1软件对获得序列进行处理。OTUs聚类采用UPARSE[12]软件以97%的相似度阈值为标准进行。使用RDP Classifier软件对OTU代表序列进行物种注释[13]。
根据Li等[14]构建的565株HPB菌株的数据库,从NCBI的GenBank中获得HPB的16S rRNA基因序列。采用BLASTN软件进行序列比对,以E值<×10-10,同一性≥ 99%为阈值。样品中HPB的相对丰度为比对得到HPB序列数与总序列数的比值。
1.5 数据处理
常规的数据处理与统计分析使用Microsoft Excel 2010完成。利用R语言的pheatmap包绘制热图。采用基于Bray-Curtis距离的主坐标分析(principal coordinate analysis, PCoA)方法分析了不同处理土壤中细菌群落结构及HPB组成差异。采用Mantel检验研究了土壤性质、细菌群落与HPB之间的相关性。采用方差分解分析(variation partitioning analysis, VPA)来评估不同因素解释的HPB变异的百分比。计算HPB和ARGs的Spearman秩相关性,并采用Cytoscape 3.8.2基于圆形布局可视化其中显著(ρ>0.5,P<0.001)的相关关系。
2 结果(Results)
2.1 土壤中细菌群落结构特征
由图1(a)可知,PCoA分析显示前两轴共解释41.6%的细菌群落结构变化,其中不同施肥处理样品在第一轴上分开,表明施加粪肥与施加不同类型粪肥能明显影响土壤细菌群落结构(PERMANOVA test, P<0.001)。由图2(a)可知,不同粪肥处理中Shannon指数表现为CK>CMs>PMs,说明施用畜禽粪便降低了紫色土中细菌群落的多样性,其中施用猪粪处理最为明显。施肥后土壤中的细菌多样性随时间呈现出先升高后降低的趋势,其中以第0天样品最低。而病原菌Shannon指数在第0天和第23天表现为PMs>CMs>CK,到第45天之后表现为PMs>CK>CMs,说明施加猪粪增加了土壤中病原菌的多样性,而施加鸡粪降低了土壤中病原菌多样性(图2(b))。
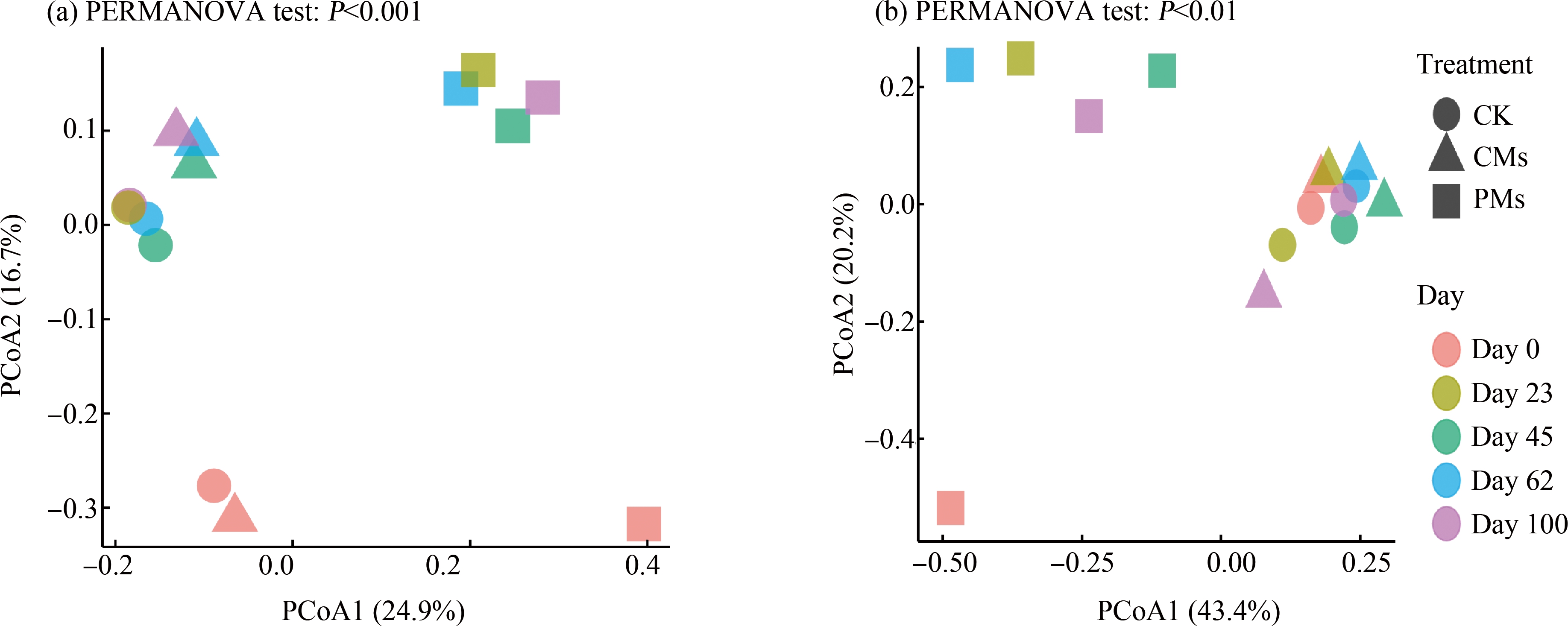
图1 未施肥土壤和施肥土壤中细菌群落(a)和人类致病菌(b)的主坐标分析
Fig. 1 PCoA ordination of bacterial communities (a) and human pathogens (b) in unfertilized and fertilized soils
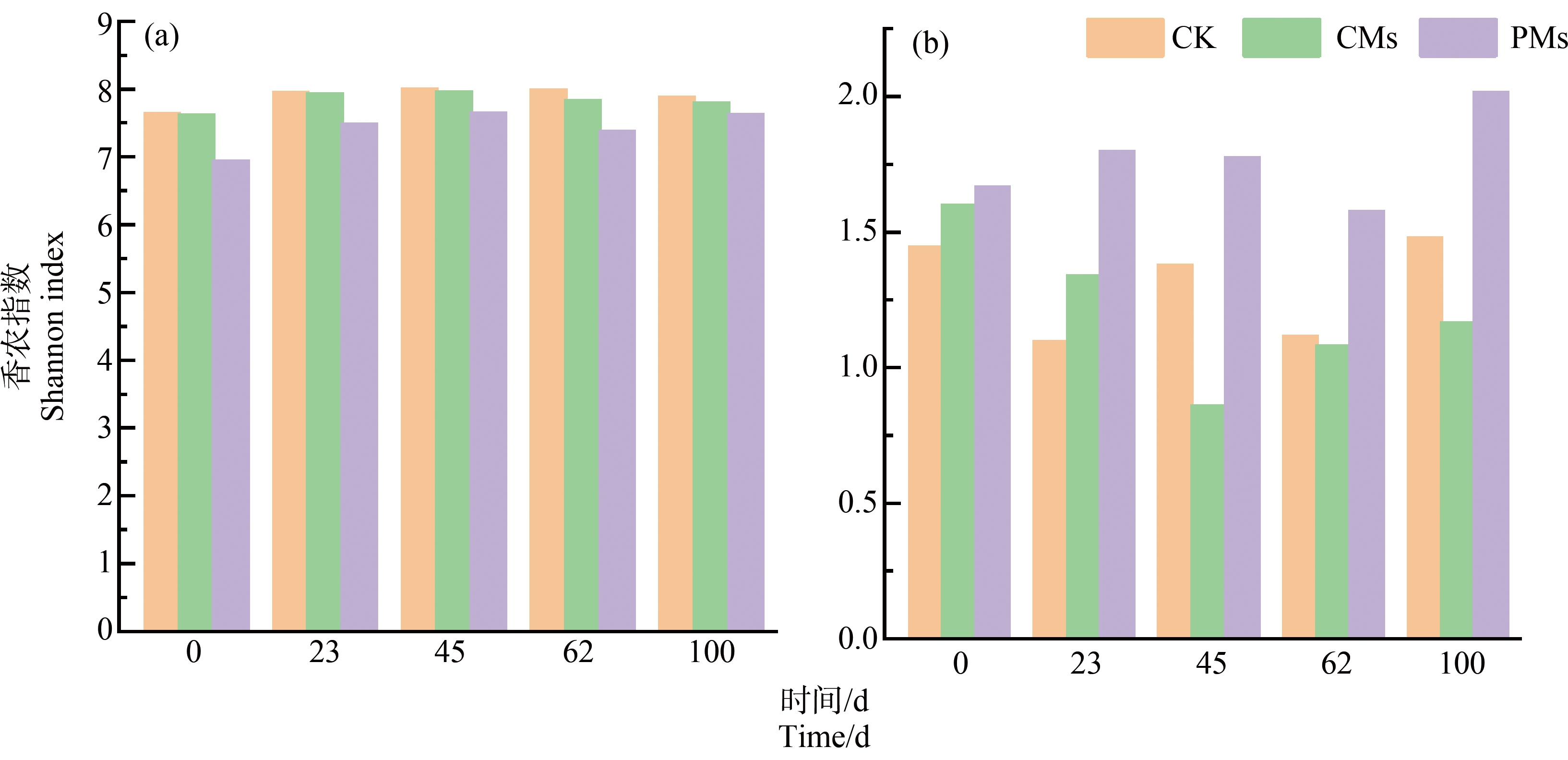
图2 不同处理土壤中微生物群落多样性(a)与病原菌多样性(b)的时间变化
Fig. 2 Temporal variation of microbial community diversity (a) and pathogen diversity (b) in soils under different treatments
2.2 土壤中致病菌的分布
在所有样本中检测到16种人类致病菌,其中猪粪、鸡粪和未施肥土壤分别检测到13种、5种和9种。由图3(a)可知,猪粪和未施肥土壤分别独有7种和3种病原菌,而鸡粪中携带的病原菌均能在猪粪和未施肥土壤中检出。由图3(b)及图4可知,在所有样本中猪粪样本的致病菌相对丰度最高,其中主要包括大肠链球菌属(Streptococcus gallolyticus strain ATCC BAA-2069)、大肠杆菌属(Escherichia_coli_APEC_O78)、猪链球菌属(Streptococcus suis BM407)和谷氨酸棒状杆菌(Corynebacterium_glutamicum_R)。同时,在培养过程中致病菌的组成发生变化。由图1(b)可知,施加猪粪处理的土壤中致病菌组成与施加鸡粪土壤、未施肥土壤有明显不同(P<0.01)。由此可见,猪粪中的致病菌丰度明显高于未施肥土壤和鸡粪,且施加猪粪会显著改变土壤中致病菌组成。
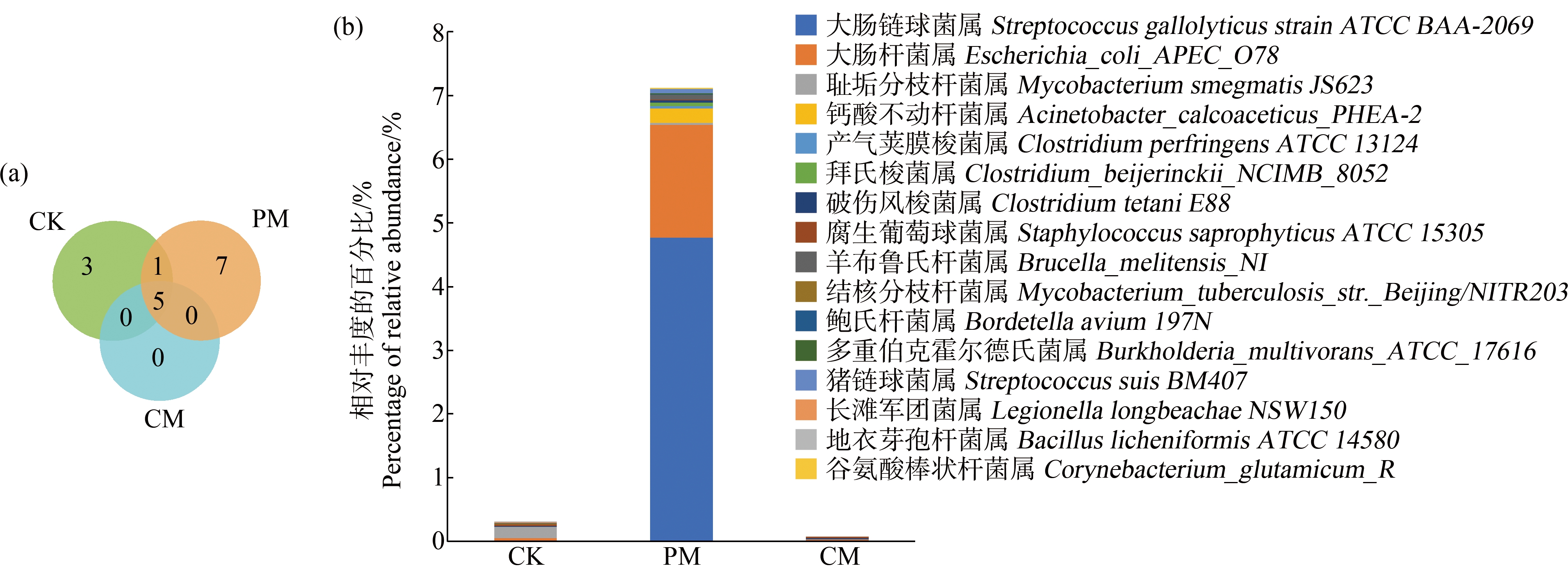
图3 未施肥土壤和粪肥中携带的共有和独有致病菌数量(a)及致病菌相对丰度(b)
注:CK表示未施肥土壤;PM表示猪粪;CM表示鸡粪。
Fig. 3 Number of shared and unique pathogenic bacteria (a) and relative abundance of pathogenic bacteria (b) in unfertilized soil and manure
Note: CK means unfertilized soil; PM means pig manure; CM means chicken manure.
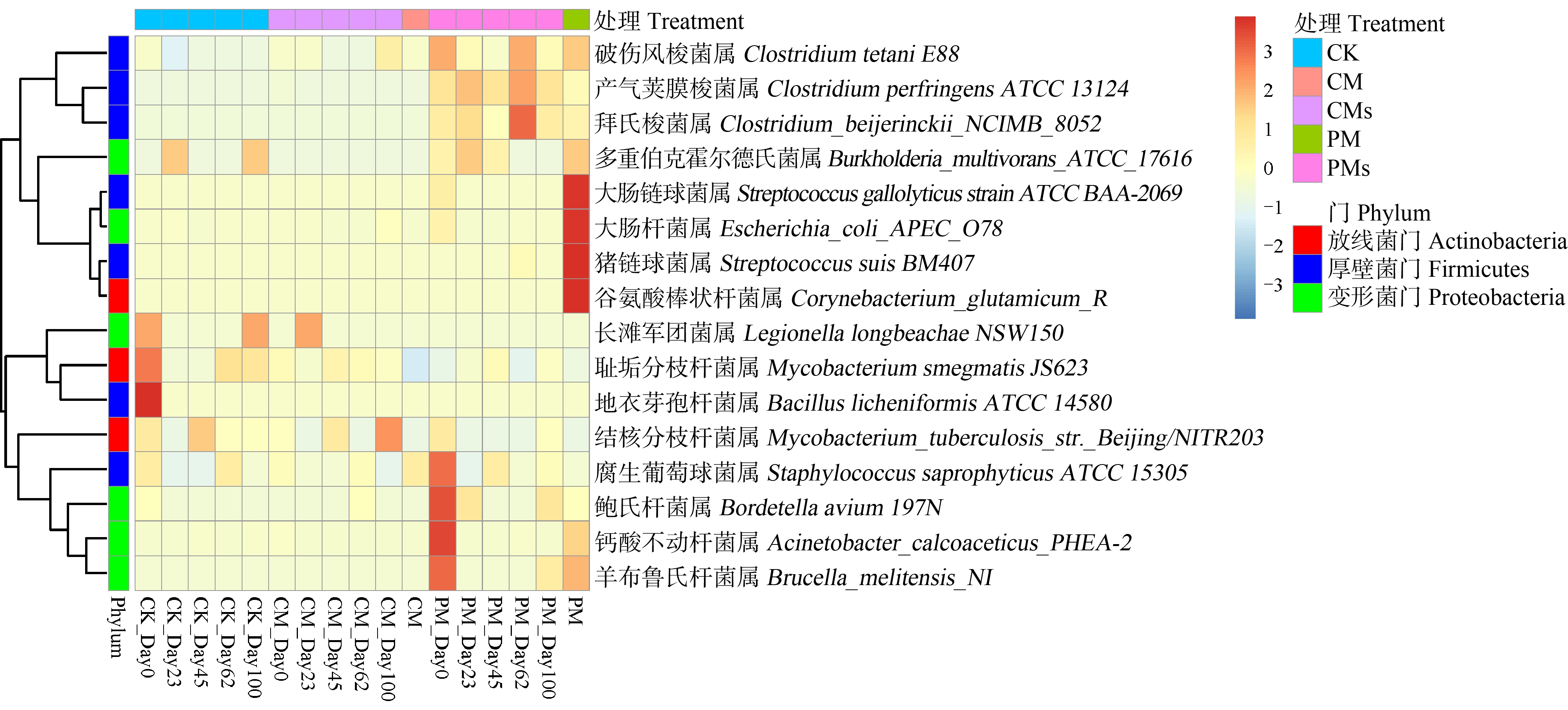
图4 不同培养时间以及不同处理中病原菌的相对丰度(%)
注:相对丰度没有进行log换算。
Fig. 4 The relative abundance of pathogens under different incubation times and treatments (%)
Note: Relative abundances were not log converted.
由图4可知,整体上看致病菌的相对丰度随培养时间而不断降低。与未施肥土壤相比,施加鸡粪对土壤致病菌的相对丰度没有明显影响。而施加猪粪显著提高了腐生葡萄球菌属(Staphylococcus saprophyticus ATCC 15305)、鲍氏杆菌属(Bordetella avium 197N)、钙酸不动杆菌属(Acinetobacter_calcoaceticus_PHEA-2)以及羊布鲁氏杆菌属(Brucella_melitensis_NI)的相对丰度。这与猪粪中致病菌丰度有关[15]。在整个培养过程中,施加猪粪后土壤中不同致病菌的相对丰度发生明显变化。培养时间到第62天,梭菌属的致病菌Clostridium_beijerinckii_NCIMB_8052、Clostridium perfringens ATCC 13124、Clostridium tetani E88相对丰度明显增加,而腐生葡萄球菌属(Staphylococcus saprophyticus ATCC 15305)、鲍氏杆菌属(Bordetella avium 197N)、钙酸不动杆菌属(Acinetobacter_calcoaceticus_PHEA-2)以及羊布鲁氏杆菌属(Brucella_melitensis_NI)相对丰度明显减少。除此之外,可以看到主要的致病菌门有3类,即放线菌门(Actinobacteria)、厚壁菌门(Firmicutes)和变形菌门(Proteobacteria)。Li等[14]研究发现猪粪中致病菌比例高于鸡粪,但施加鸡粪对土壤中致病菌相对丰度的改变比施加猪粪更明显。与之不同的是,也有研究指出施加猪粪土壤中致病菌相对丰度高于施加鸡粪土壤[16],结果的差异可能是因为粪便样本不同。
2.3 ARGs和HPB之间的共现关系网络
土壤中ARGs的数量、丰度和组成变化已在笔者先前的研究中报道[17]。由图5可知,该网络由55个节点(8个HPB和47个ARGs)和79条边构成。8种HPB中拜氏梭菌属(Clostridium_beijerinckii_NCIMB_8052)、产气荚膜梭菌属(Clostridium perfringens ATCC 13124)和羊氏布鲁杆菌属(Brucella_melitensis_NI)与多个ARGs呈显著正相关关系。Fang等[18]通过宏基因组测序发现大多数ARGs可以通过基因水平迁移在致病菌之间转移,并且提出与ARGs共现的HPB可视之为ARGs的宿主菌。由图5可知,产气荚膜梭菌属(Clostridium perfringens ATCC 13124)可能是27种ARGs的潜在宿主,而拜氏梭菌属(Clostridium_beijerinckii_NCIMB_8052)可能作为33种ARGs的潜在宿主,羊氏布鲁杆菌属(Brucella_melitensis_NI)可能作为6种ARGs的潜在宿主。

图5 基于Spearman相关系数抗生素抗性基因(ARGs)与测定的人类致病菌(HPB)之间的共现关系(ρ>0.8, P<0.001)
注:不同颜色的四边形和圆形节点分别代表HPB所属门类和ARGs产生耐药性的抗生素。
Fig. 5 The network graph showing the co-occurring patterns among antibiotic resistance genes (ARGs) and detected human pathogen bacteria (HPB) based on Spearman’s correlation (ρ>0.8, P<0.001)
Note: Quadrilateral and circular nodes in different colors represent the class of antibiotics that HPB belongs to and the antibiotics that ARGs has developed resistance to, respectively.
2.4 影响培养期间HPB相对丰度的因素
Mantel分析表明,HPB相对丰度与细菌群落结构有显著的相关性(r20.44,P<0.01),与土壤SOC含量有显著关系(r2=0.25,P<0.05),与TN含量没有显著关系(r2=0.22,P>0.05)。VPA分析表明,土壤理化性质和细菌群落结构对HPB相对丰度变化具有70.6%的解释量,说明两者是影响土壤中HPB相对丰度的主要因素(图6)。相似地,Li等[14]研究发现HPB的主要预测因子是土壤细菌群落结构与理化性质的变化。
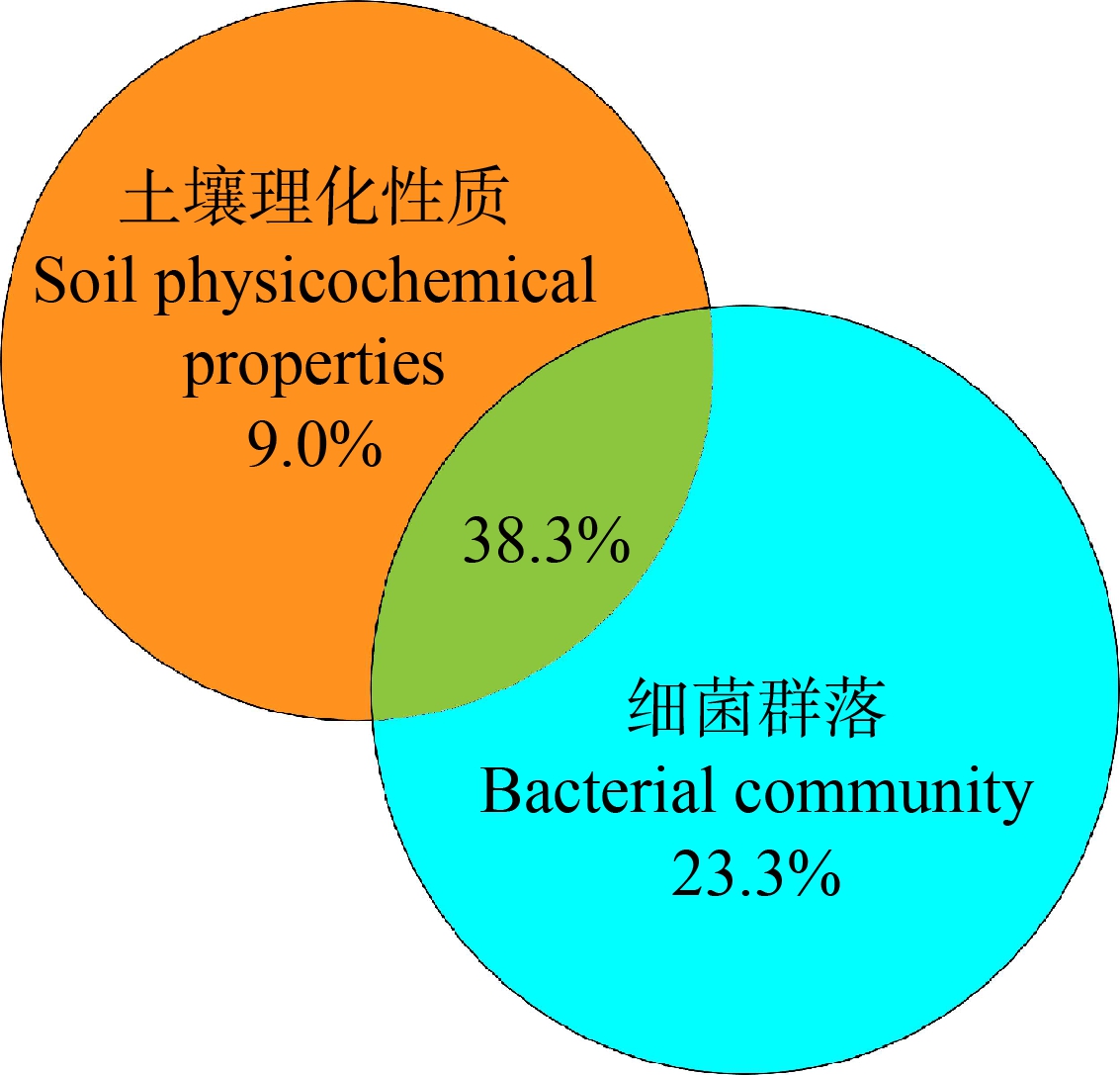
图6 土壤理化性质和细菌群落结构对HPB的变差解释百分比(VPA分析)
Fig. 6 The percentage of HPB variation explained by soil physio-chemical properties and bacterial communities (VPA analysis)
3 讨论(Discussion)
3.1 施加粪肥对土壤微生物群落及致病菌分布的影响
微生物群落在土壤生化过程中具有关键性作用。施加粪肥可以增加土壤中的土壤有机碳等养分[19],从而提高微生物生物量改变土壤微生物群落[20]。林曼霞等[21]研究发现抗生素残留会影响施加粪肥土壤中反硝化作用,改变菌群丰度。研究发现粪便中携带的抗生素会对粪便细菌与土壤细菌产生选择性压力,导致细菌群落结构改变[22]。本研究表明施加粪肥处理土壤的细菌群落组成与对照处理有明显区别,这与前人的研究结果一致[23]。此外,不同粪肥处理对土壤微生物群落的影响也存在差异,如安娜等[24]研究发现施用牛粪和鹿粪的土壤中生防真菌支顶孢属和青霉菌属丰度明显升高,而施加鸡粪的土壤中丰度显著升高的是病原菌镰刀菌属。Pérez-Valera等[25]发现施加牛粪会改变土壤细菌群落结构,并引发不动杆菌在土壤中的短期优势。畜禽粪便含的丰富致病菌[26],可在粪肥土地利用过程中进入土壤并在土壤环境中完成定殖[27-28]。理鹏等[29]研究发现土壤添加鸡粪或猪粪均会导致病原菌相对丰度增加,其中施用鸡粪和猪粪分别显著增加了假单胞菌属和黄杆菌属致病菌的相对丰度。Li等[30]的研究结果表明施加粪肥显著增加了表层土壤中的病原菌丰度,同时还发现TOC能明显影响HPB分布。本研究结果表明施加粪肥后土壤中的致病菌组成发生变化,致病菌丰度随着时间不断下降,推测是由于土壤的自动净化机制、粪源致病菌的竞争力因资源限制而降低及其他影响[25,31]。致病菌丰度的大幅度下降说明这类致病菌在与实验土壤相似的条件下生存能力很低[32]。营养供应是施加粪肥提高土壤肥力的主要途径,也是影响微生物生存的重要因素。固有微生物得到营养供应后,竞争力快速提升,生存能力增强[33]。研究也表明病原菌的持久性与是否持续施肥有关,如果不再施加粪肥,土壤中的病原菌丰度会随时间下降[34]。
3.2 致病菌与ARGs的关系
已有研究表明畜禽粪便污染的土壤是病原菌和抗生素耐性的储存库[35]。多项研究表明多药耐药基因是ARGs最丰富的种类[36-37],同时一种病原菌可以携带多个ARGs,因此携带ARGs的致病菌极有可能具有多种抗生素耐性。本研究中发现施加粪肥的土壤中存在大量病原菌与ARGs,并且一种病原菌与多个ARGs之间存在显著相关关系。证明施肥土壤中的潜在病原菌可能具有多重耐药性。与前人研究一致,邓雯文等[38]研究发现粪肥中潜在致病菌与抗性基因tetM及sul1呈显著正相关。此外,土壤中的潜在致病菌还可以携带ARGs向植物迁移或残留在土壤中,从而危害食物的健康安全[30,37,39]。
3.3 施肥土壤中致病菌的影响因素
本研究中,土壤性质和细菌群落分别单独解释了9.0%和23.3%的HPB变化。土壤细菌群落结构的较高贡献可能与土著微生物对粪源病原菌生长的抑制作用有关。Goberna等[5]研究发现潜在病原菌在γ辐射后土壤中的数量比在未辐射处理中更丰富,证明了土著微生物对潜在病原菌生长的抑制。值得注意的是,土壤性质和细菌群落的相互作用可以解释38.3%的变化,高于它们单独的解释量。Moynihan等[28]研究发现不同土地利用方式的土壤微生物群落结构与理化性质各不相同,二者的相互作用对土壤中病原体存活起到重要调控作用。关于影响HPB变化的其他因素还需要进一步探索。
[1] 仇焕广, 廖绍攀, 井月, 等. 我国畜禽粪便污染的区域差异与发展趋势分析[J]. 环境科学, 2013, 34(7): 2766-2774
Qiu H G, Liao S P, Jing Y, et al. Regional differences and development tendency of livestock manure pollution in China [J]. Environmental Science, 2013, 34(7): 2766-2774 (in Chinese)
[2] 刘春, 刘晨阳, 王济民, 等. 我国畜禽粪便资源化利用现状与对策建议[J]. 中国农业资源与区划, 2021, 42(2): 35-43
Liu C, Liu C Y, Wang J M, et al. Thecurrent situation of resource utilization of livestock and poultry manure in China and the countermeasures and suggestions [J]. Chinese Journal of Agricultural Resources and Regional Planning, 2021, 42(2): 35-43 (in Chinese)
[3] 张克强, 杜连柱, 杜会英, 等. 国内外畜禽养殖粪肥还田利用研究进展[J]. 农业环境科学学报, 2021, 40(11): 2472-2481, 2591
Zhang K Q, Du L Z, Du H Y, et al. Application of livestock and poultry waste to agricultural land: A review [J]. Journal of Agro-Environment Science, 2021, 40(11): 2472-2481, 2591 (in Chinese)
[4] Liu S B, Wang J Y, Pu S Y, et al. Impact of manure on soil biochemical properties: A global synthesis [J]. The Science of the Total Environment, 2020, 745: 141003
[5] Goberna M, Podmirseg S M, Waldhuber S, et al. Pathogenic bacteria and mineral N in soils following the land spreading of biogas digestates and fresh manure [J]. Applied Soil Ecology, 2011, 49: 18-25
[6] Heinonen-Tanski H, Mohaibes M, Karinen P, et al. Methods to reduce pathogen microorganisms in manure [J]. Livestock Science, 2006, 102(3): 248-255
[7] van Bruggen A H C, Goss E M, Havelaar A, et al. One Health—Cycling of diverse microbial communities as a connecting force for soil, plant, animal, human and ecosystem health [J]. Science of the Total Environment, 2019, 664: 927-937
[8] Fu S Z, Wang Q Y, Wang R, et al. Horizontal transfer of antibiotic resistance genes within the bacterial communities in aquacultural environment [J]. Science of the Total Environment, 2022, 820: 153286
[9] Udikovic-Kolic N, Wichmann F, Broderick N A, et al. Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization [J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(42): 15202-15207
[10] Forsberg K J, Reyes A, Wang B, et al. The shared antibiotic resistome of soil bacteria and human pathogens [J]. Science, 2012, 337(6098): 1107-1111
[11] Tamaki H, Wright C L, Li X Z, et al. Analysis of 16S rRNA amplicon sequencing options on the Roche/454 next-generation titanium sequencing platform [J]. PLoS One, 2011, 6(9): e25263
[12] Edgar R C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads [J]. Nature Methods, 2013, 10(10): 996-998
[13] Wang Q, Garrity G M, Tiedje J M, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy [J]. Applied and Environmental Microbiology, 2007, 73(16): 5261-5267
[14] Li J Y, Chen Q L, Li H L, et al. Impacts of different sources of animal manures on dissemination of human pathogenic bacteria in agricultural soils [J]. Environmental Pollution, 2020, 266(Pt 2): 115399
[15] McCarthy G, Lawlor P G, Gutierrez M, et al. Assessing the biosafety risks of pig manure for use as a feedstock for composting [J]. The Science of the Total Environment, 2013, 463-464: 712-719
[16] Zhu L, Lian Y L, Lin D, et al. Insights into microbial contamination in multi-type manure-amended soils: The profile of human bacterial pathogens, virulence factor genes and antibiotic resistance genes [J]. Journal of Hazardous Materials, 2022, 437: 129356
[17] Cheng J H, Tang X Y, Liu C. Bacterial communities regulate temporal variations of the antibiotic resistome in soil following manure amendment [J]. Environmental Science and Pollution Research, 2021, 28(23): 29241-29252
[18] Fang H, Han L X, Zhang H P, et al. Dissemination of antibiotic resistance genes and human pathogenic bacteria from a pig feedlot to the surrounding stream and agricultural soils [J]. Journal of Hazardous Materials, 2018, 357: 53-62
[19] Ozlu E, Kumar S. Response of soil organic carbon, pH, electrical conductivity, and water stable aggregates to long-term annual manure and inorganic fertilizer [J]. Soil Science Society of America Journal, 2018, 82(5): 1243-1251
[20] Gautam A, Sekaran U, Guzman J, et al. Responses of soil microbial community structure and enzymatic activities to long-term application of mineral fertilizer and beef manure [J]. Environmental and Sustainability Indicators, 2020, 8: 100073
[21] 林曼霞, 邹勇, 孙永学. 室内粪土模型中土霉素对主要反硝化基因转录水平和菌群结构特征的影响[J]. 生态毒理学报, 2018, 13(5): 182-189
Lin M X, Zou Y, Sun Y X. Effects of oxytetracycline on the transcriptional characteristics of dominant denitrification genes and flora structure characteristics in indoor manure-soil model [J]. Asian Journal of Ecotoxicology, 2018, 13(5): 182-189 (in Chinese)
[22] 隋倩雯, 张俊亚, 魏源送, 等. 畜禽养殖过程抗生素使用与耐药病原菌及其抗性基因赋存的研究进展[J]. 生态毒理学报, 2015, 10(5): 20-34
Sui Q W, Zhang J Y, Wei Y S, et al. Veterinary antibiotics use, occurrence of antibiotic resistance pathogen and its antibiotic resistance genes in animal production: An overview [J]. Asian Journal of Ecotoxicology, 2015, 10(5): 20-34 (in Chinese)
[23] Shawver S, Wepking C, Ishii S, et al. Application of manure from cattle administered antibiotics has sustained multi-year impacts on soil resistome and microbial community structure [J]. Soil Biology and Biochemistry, 2021, 157: 108252
[24] 安娜, 高纪超, 韩雅棋, 等. 施粪肥对人参栽培土壤理化性质和真菌群落结构的影响[J]. 吉林农业大学学报, 2019, 41(6): 695-706
An N, Gao J C, Han Y Q, et al. Effects of manure application on soil physicochemical properties and fungal community structure in ginseng-planted soil [J]. Journal of Jilin Agricultural University, 2019, 41(6): 695-706 (in Chinese)
[25] Pérez-Valera E, de Melo Rangel W, Elhottová D. Cattle manure application triggers short-term dominance of Acinetobacter in soil microbial communities [J]. Applied Soil Ecology, 2022, 176: 104466
[26] Tulayakul P, Boonsoongnern A, Kasemsuwan S, et al. Comparative study of heavy metal and pathogenic bacterial contamination in sludge and manure in biogas and non-biogas swine farms [J]. Journal of Environmental Sciences (China), 2011, 23(6): 991-997
[27] Hu Y A, Cheng H F, Tao S. Environmental and human health challenges of industrial livestock and poultry farming in China and their mitigation [J]. Environment International, 2017, 107: 111-130
[28] Moynihan E L, Richards K G, Brennan F P, et al. Enteropathogen survival in soil from different land-uses is predominantly regulated by microbial community composition [J]. Applied Soil Ecology, 2015, 89: 76-84
[29] 理鹏, 吴建强, 沙晨燕, 等. 粪肥和有机肥施用对稻田土壤微生物群落多样性影响[J]. 环境科学, 2020, 41(9): 4262-4272
Li P, Wu J Q, Sha C Y, et al. Effects of manure and organic fertilizer application on soil microbial community diversity in paddy fields [J]. Environmental Science, 2020, 41(9): 4262-4272 (in Chinese)
[30] Li H Y, Zheng X Q, Tan L, et al. The vertical migration of antibiotic-resistant genes and pathogens in soil and vegetables after the application of different fertilizers [J]. Environmental Research, 2022, 203: 111884
[31] Chen Q L, An X L, Li H, et al. Do manure-borne or indigenous soil microorganisms influence the spread of antibiotic resistance genes in manured soil? [J]. Soil Biology and Biochemistry, 2017, 114: 229-237
[32] Pérez-Valera E, Kyselková M, Ahmed E, et al. Native soil microorganisms hinder the soil enrichment with antibiotic resistance genes following manure applications [J]. Scientific Reports, 2019, 9(1): 6760
[33] Macedo G, van Veelen H P J, Hernandez-Leal L, et al. Targeted metagenomics reveals inferior resilience of farm soil resistome compared to soil microbiome after manure application [J]. The Science of the Total Environment, 2021, 770: 145399
[34] Zhang H P, Zhang Q K, Song J J, et al. Tracking resistomes, virulence genes, and bacterial pathogens in long-term manure-amended greenhouse soils [J]. Journal of Hazardous Materials, 2020, 396: 122618
[35] Mills M, Lee S, Evans M, et al. Enteric pathogens and carbapenem resistance genes are widespread in the fecal contaminated soils of cattle farms in the United States [J]. Environmental Advances, 2021, 6: 100137
[36] Zhou R J, Zeng S Z, Hou D W, et al. Occurrence of human pathogenic bacteria carrying antibiotic resistance genes revealed by metagenomic approach: A case study from an aquatic environment [J]. Journal of Environmental Sciences (China), 2019, 80: 248-256
[37] Yin Y, Zhu D, Yang G, et al. Diverse antibiotic resistance genes and potential pathogens inhabit in the phyllosphere of fresh vegetables [J]. The Science of the Total Environment, 2022, 815: 152851
[38] 邓雯文, 杨盛智, 何雪萍, 等. 牛粪发酵过程中抗生素耐药基因及相关菌群组成变化规律[J]. 生态毒理学报, 2019, 14(2): 153-163
Deng W W, Yang S Z, He X P, et al. Change of antibiotic resistance genes and bacterial communities during dairy manure composting process [J]. Asian Journal of Ecotoxicology, 2019, 14(2): 153-163 (in Chinese)
[39] Wang F H, Sun R B, Hu H W, et al. The overlap of soil and vegetable microbes drives the transfer of antibiotic resistance genes from manure-amended soil to vegetables [J]. The Science of the Total Environment, 2022, 828: 154463