-
大气中温室气体浓度持续升高是导致全球气候变暖的主要原因[1]。甲烷 (CH4) 作为第二重要的温室气体,其全球变暖潜力是二氧化碳 (CO2) 的28~34倍[2]。畜禽养殖业是大气中CH4重要的人为排放源,约占全球人为CH4排放量的28%~35%[3],因此,控制畜禽养殖业CH4排放对于削减人为源温室气体排放至关重要。畜禽养殖业排放的CH4约11%来自粪尿存储过程微生物的厌氧发酵[4]。近些年,畜禽粪尿由敞开式存储转变为密闭式存储成为主流趋势[5],这使得采取工程措施对粪尿存储产生的含CH4废气进行控制成为可能。与沼气池不同,粪尿存储过程的厌氧发酵活动水平一般不高,这导致粪尿存储排放的含CH4废气浓度一般不会超过5%,不具备能源化利用价值,也不宜采用建设和运行成本均较高的吸附法或催化燃烧法进行处理[6]。
生物过滤废气净化技术具有建设成本低、运行维护简单等优点,适合于处理养殖场排放的低浓度、大气量含CH4废气[7]。生物过滤净化CH4过程是在一定的人工控制条件下,利用甲烷氧化菌将CH4转化为CO2和水的微生物学过程[8]。上述人工控制条件主要包括营养物质[9]、CH4/O2[10]、pH[11]、温度[12]、水分[13]、空床停留时间[14]等,这其中以氮源为代表的营养物质供给对生物过滤塔CH4净化效率的影响最为关键。只有持续的氮源供应才能保证生物过滤塔高效稳定的CH4净化性能。因此,生物过滤法常采用堆肥作为滤料,堆肥中养分的缓慢释放可供给甲烷氧化菌所需的含氮营养物质。然而,当堆肥中营养物质耗尽后,生物过滤塔的CH4氧化速率则会降低或者停止,需频繁更换滤料来维持运行[15]。为避免上述滤料更换问题,目前常采用在滤料上循环喷淋营养液的形式来补充生物过滤塔中甲烷氧化菌所需的氮源等营养物质,但这会增大技术的运行成本[16],因此,寻求一种价格低廉、来源广泛的液态氮源对于加快生物法净化CH4废气技术的应用显得尤为重要。
畜禽养殖场液态粪污一般含有高浓度的铵态氮[17]。这类废水是否可作为除CH4生物过滤塔的营养物质,目前尚未给出清晰答案。一般而言,硝态氮被认为是最适宜甲烷氧化菌生长代谢的营养物质。如以硝态氮为主要氮源的硝酸盐无机培养基 (NMS培养基) 就被广泛用于甲烷氧化纯菌的培养[18]。NIKIEMA等[19]以硝态氮作为生物过滤法净化CH4废气的氮源,使得CH4的去除负荷高达36 g·m−3·h−1。相对而言,铵态氮常被认为其对甲烷氧化菌具有抑制作用,其原因主要有:一是氨单加氧酶 (AMO) 和甲烷单加氧酶 (pMMO) 从进化亲缘关系上属于同一类含铜的单加氧酶,铵态氮会与CH4竞争pMMO上的活性位点,进而抑制CH4氧化过程;二是铵氮转化过程中会积累一定浓度的NH2OH和NO2−,这2种物质对微生物具有毒性作用,会长期抑制甲烷氧化菌的生长代谢[20]。如HUBER等[21]发现高浓的铵态氮会抑制甲烷氧化纯菌的活性,从而降低生物过滤床的CH4净化效率。但是,NYERGES等[22]研究表明,甲烷氧化菌Methylomicrobium album在铵态氮较高的环境中具有高于其他甲烷养化菌的竞争优势,推测某些甲烷氧化菌可与其他转化氨的微生物组成一个共生群落,进而维持微型生态系统的平衡。该研究结果为含高浓度铵态氮的液态粪污作为生物过滤法净化CH4废气的氮源提供了可能。为探明铵态氮作为氮源对生物过滤塔CH4净化过程的影响,还应对该过程涉及到的微生物学机理进行深入研究。
本研究以不加氮源生物过滤塔为对照,开展铵态氮对生物过滤塔CH4净化性能的影响,并借助宏基因组学分析技术,从物种分类学组成、系统代谢功能方面,解析铵态氮对生物过滤塔净化CH4过程微生物群落结构和功能的影响机制,以期为生物过滤法净化畜禽养殖含CH4废气和液态养殖粪污的综合利用提供参考。
-
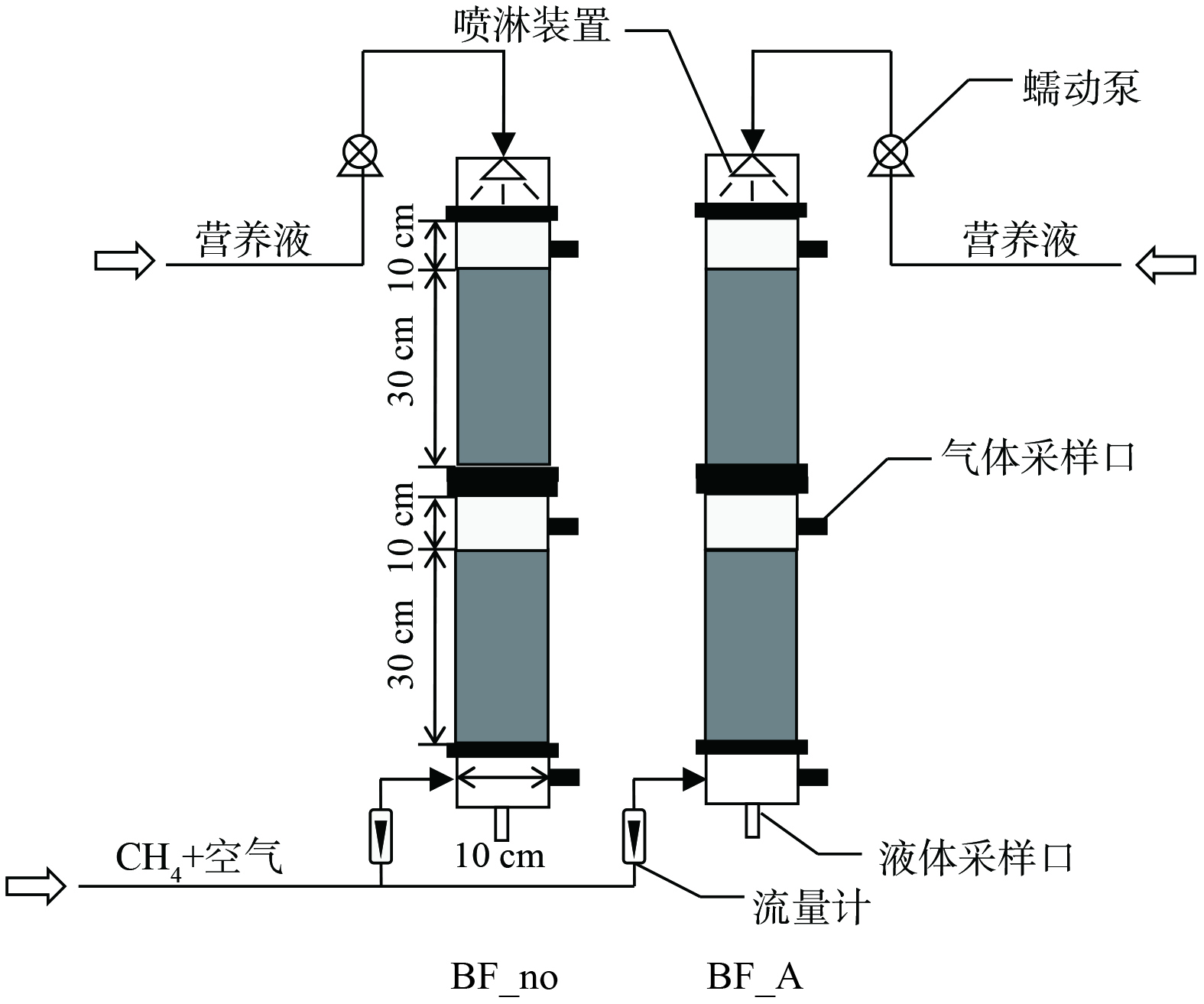
本研究共运行2个生物过滤塔,分别命名为BF_A和BF_no (图1) ,其中生物过滤塔BF_A供给铵态氮氮源,其营养液成分为MgSO4·7H2O 1.00 g;NH4Cl 0.53 g;Na2HPO4·12H2O 0.72 g;KH2PO4 0.27 g;CaCl2·6H2O 0.20 g;EDTA 铁氨 5 mg;蒸馏水1 L[23]。对照生物过滤塔BF_no不供给氮源,其他营养液成分同生物过滤塔BF_A。2个生物过滤塔均采用有机玻璃制成,分为上、下2层,每层高40 cm,内径为10 cm,壁厚0.5 cm。生物过滤塔滤料由垃圾填埋场覆土与活性炭 (2~5 mm) 按3∶1混合制成,每层滤料高30 cm,总体积为4.71 L。在生物过滤塔的底部和顶部各设置1个气体采样口,采用医用注射器采集气体样品并打入气相色谱仪分析CH4浓度。实验用气为模拟养殖场含CH4废气,由钢瓶气减压后与压缩空气混合而成,废气由生物过滤塔底部进入生物过滤塔,经填料层被微生物净化后,由生物过滤塔顶部出气口排出。研究期间,生物过滤塔的运行条件为:进气CH4质量浓度1 500~5 000 mg·m−3,气体流量0.03~0.3 L·min−1。生物过滤塔采用间歇式营养液供给模式,每3 d喷淋1次,每次250 mL,营养液由蠕动泵输送至生物过滤塔,从顶部喷淋至填料表层,与含CH4废气逆流接触,最终从过滤塔底部流出。
-
1) 样品采集。在生物过滤塔成功挂膜启动后,在生物过滤塔空床停留时间 (Empty bed residence time,EBRT) 为56 min且稳定运行2 d后,分别从2个生物过滤塔上、下层各提取25 g生物填料,每个生物过滤塔的生物填料均匀混合为1个样品中,每个样品3次重复,置于−80 ℃冰箱中保存,用于DNA的提取。
2) 基因组提取。利用E.Z.N.A.® Soil DNA Kit (Omega Bio-tek,美国) 试剂盒进行生物填料样品DNA抽提。基因组DNA抽提完成后,利用TBS-380检测DNA浓度,利用NanoDrop200检测DNA纯度,利用1%琼脂糖凝胶电泳检测DNA的完整性。通过Covaris M220仪器将DNA片段化,筛选约400bp的片段,使用NEXTflexTM Rapid DNA-Seq (Bioo Scientific,美国) 构建PE文库,并使用Illumina NovaSeq/Hiseq Xten (Illumina,美国) 测序平台进行宏基因组测序 (上海美吉生物医药科技有限公司) 。
3) 生物信息学分析 采用fastp[24] (https://github.com/OpenGene/fastp,version 0.20.0) 对原始测序数据进行质量控制,质量控制后获得的短片段序列采用软件MEGAHIT[25] (https://github.com/voutcn/megahit,version 1.1.2) 进行进行拼接组装。在拼接结果中筛选≥300 bp的contigs作为最终的组装结果。随后,使用MetaGene[26] (http://metagene.cb.k.u-tokyo.ac.jp/) 对拼接结果中的contigs进行ORF预测,选择核酸长度大于等于100bp的基因,并将其翻译为氨基酸序列。获得基因预测结果后,采用CD-HIT[27] (http://www.bioinformatics.org/cd-hit/,version 4.6.1) 对所有样品预测出来的基因序列进行聚类 (参数为:90% identity、90% coverage) ,每类取最长的基因作为代表序列,构建非冗余基因集。而后,使用SOAPaligner[28]软件 (http://soap.genomics.org.cn/,version 2.21) ,分别将每个样品的高质量reads与非冗余基因集进行比对 (95% identity) ,统计基因在对应样品中的丰度信息。最后,使用Diamond[29] (http://www.diamondsearch.org/index.php,version 0.8.35) 将非冗余基因集的氨基酸序列分别与NR数据库和KEGG数据库 (version 94.2) 进行比对 (BLASTP比对参数设置期望值e-value为1e-5) ,以此获得物种注释和基因对应的KEGG功能。
-
生物过滤塔进、出气中的CH4浓度采用气相色谱仪 (Agilent 6820) 进行测定。测定条件为:以N2为载气,ECD检测器,柱箱温度为55 ℃,前检测器温度为250 ℃,后检测器温度为330 ℃。
-
进气CH4质量浓度维持在1 500~2 500 mg·m−3,EBRT为131 min,通过分析2个生物过滤塔进、出口CH4浓度,比较2个生物过滤塔的挂膜启动性能。图2表明,2个生物过滤塔在挂膜初期的CH4净化效率约为10%,从第8天开始迅速升高,2个生物过滤塔的CH4去除效率在第18天均达到了80%以上,整个挂膜启动共历时约27 d。2个生物过滤塔的CH4去除效率并没有显著差异,推断原因主要为2个生物过滤塔运行的EBRT较长,生物过滤塔中微生物具备充足时间完成CH4氧化过程,因此,2个生物过滤塔在挂膜启动阶段的CH4净化效率不存在明显差异。
-
停留时间 (EBRT) 是生物过滤塔运行的一个重要参数,适当延长EBRT有助于提高生物过滤塔CH4去除效率,但EBRT过长会导致生物过滤塔占地面积过大,增加建设和运行成本,限制该技术的工业应用[30]。在进气CH4质量浓度为4 000~5 000 mg·m−3的运行条件下,考察了不同EBRT (131 min、78 min、56 min、44 min、32 min) 对2个生物过滤塔CH4去除效率的影响,每个停留时间运行7 d。由图3可知,当EBRT为78和131 min时,2个生物过滤塔均能够保持约90%的CH4去除效率,其中以铵态氮为氮源的BF_A要高于对照生物过滤塔BF_no。然而,当生物过滤塔的EBRT缩短为56和44 min时,生物过滤塔BF_A仍能够维持较高的CH4去除效率 (84%~88%) ,显著高于BF_no,其CH4去除效率只有62%~74%。当EBRT缩短为32 min时,生物过滤塔BF_A的的CH4去除效率下降至约80%,但仍高于对照生物过滤塔,此时BF_no的CH4去除效率则是下降到60%以下。综上所述,以铵态氮为氮源的生物过滤塔具有更优异的CH4去除性能。
-
生物过滤塔BF_no和BF_A的微生物样品分别获得1.57×107和1.03×108条原始序列,以及2.36×109和1.56×1010条原始碱基。经过质量控制后,原始序列和碱基均保留了96%以上,这表明本次测序有效地获取了样本的遗传信息,保证了后续分析结果的准确性和可靠性。
生物过滤塔BF_no和BF_A在门水平上的微生物群落组成如图4 (a) 所示,2个生物过滤塔的物种组成存在明显差异。变形菌门 (Proteobacteria) 为生物过滤塔BF_no和BF_A的第一优势门,占比分别为44.2%和50.1%。在对照生物过滤塔BF_no中,放线菌门 (Actinobacteria) 和芽单胞菌门 (Gemmatimonadetes) 的相对丰度比BF_A高,分别为8.2%和6.3%。而生物过滤塔BF_A中的绿弯菌门 (Chloroflexi) 占比为7.8%,高于对照生物过滤塔BF_no (5.8%) 。此外,硝化螺旋菌门 (Nitrospirae) 是生物过滤塔BF_A中的特有菌门,其相对丰度为3.3%。
在属水平上,生物过滤塔BF_no和BF_A的微生物群落组成如图4 (b) 所示。比较分析有确切分类信息的菌属可以看出,生物过滤塔BF_no和BF_A中第一优势属均为甲基杆菌属 (Methylobacter) ,其所占比例分别为2.2%和3.9%。甲基微菌属 (Methylomicrobium) 、锰硝石菌属 (Candidatus_Manganitrophus) 和硝化螺旋菌属 (Nitrospira) 是生物过滤塔BF_A中的特有菌种,占比分别为2.8%、1.7%和1.2%。硝化螺旋菌门和硝化螺旋菌属的微生物是环境中转化铵氮的主要菌属,可实现铵态氮向硝态氮的转化,硝态氮的生成可以保证甲烷氧化菌对氮源的需求。此外,还可以解除铵态氮对甲烷氧化菌的抑制作用。甲基微菌属 (Methylomicrobium) 是甲烷氧化菌的一种,与其他甲烷氧化菌相比,其生长速度更快,对环境的适应能力更强,被认为是甲烷氧化过程的“生物催化剂”[31]。锰硝石菌属 (Candidatus_Manganitrophus) 对甲烷代谢的影响尚不清楚,需进一步深入开展研究。此外,甲基暖菌属 (Methlocaldum) 在对照生物过滤塔BF_no中的相对丰度较高,占比为1.5%。值得注意的是,2个生物过滤塔均有约35.0%的物种在属水平下没有明确的分类信息,主要包括unclassified_p__Acidobacteria、unclassified_c__Gammaproteobacteria、unclassified_p__Chloroflexi、unclassified_c__Betaproteobacteria和unclassified_p__Planctomycetes等。
-
生物过滤塔BF_no和BF_A的微生物组成差异如图5所示。在门水平上,生物过滤塔BF_A中变形菌门 (Proteobacteria) 、绿弯菌门 (Chloroflexi) 、酸杆菌门 (Acidobacteria) 和硝化螺旋菌门 (Nitrospirae) 的相对丰度分别为50.1%、7.8%、5.3%和3.3%,极显著高于对照生物过滤塔BF_no (44.2%、5.8%、5.0%和0.8%) (P≤0.001) ,而放线菌门 (Actinobacteria) 、浮霉菌门 (Planctomycetes) 和疣微菌门 (Verrucomicrobia) 则主要分布在无外加氮源的对照生物过滤塔BF_no中,相对丰度分别为8.2%、6.5%和4.9%。上述结果与2个生物过滤塔的微生物群落结构对比分析结果相一致。在属水平上,生物过滤塔BF_A中甲基杆菌属 (Methylobacter) 、甲基微菌属 (Methylomicrobium) 、锰硝石菌属 (Candidatus_Manganitrophus) 、硝化螺旋菌属 (Nitrospira) 和甲基八叠球菌属 (Methylosarcina) 的相对丰度分别为3.9%、2.8%、1.7%、1.2%和1.2%,极显著高于对照生物过滤塔BF_no (2.1%、0.8%、0.0%、0.5%和0.4%) (P≤0.001),而甲基暖菌属 (Methlocaldum)、长微菌属 (Longimicrobium)、姜氏菌属 (Jiangella) 和鞘氨醇单胞菌属 (Sphingomonas) 则主要分布在对照生物过滤塔BF_no中,相对丰度分别为1.5%、1.6%、1.3%和1.1%。
-
1) 对甲烷代谢的影响。为更好地解析铵态氮对生物过滤塔中甲烷代谢的影响,基于宏基因组数据进一步分析了2个生物过滤塔中的CH4代谢路径特征,绘制的CH4代谢路径见图6 (a) 。2个生物过滤塔显示了完整的CH4代谢通路。此外,将2个生物过滤塔中的CH4代谢关键酶的基因含量进行差异性比较,结果如图6 (b) 所示。这些基因包括:编码甲烷单加氧酶的基因mmoX (EC 1.14.13.25) 和pmoA (EC 1.14.18.3) ;编码甲醇脱氢酶的基因mxaF (EC 1.1.2.7) ;编码甲醛脱氢酶的基因fdhA (EC 1.2.1.46) ;编码甲酸脱氢酶的基因FDH (EC 1.17.1.9) ;编码5,6,7,8-四氢甲蝶呤水解酶的基因fae (EC 4.2.1.147) ;编码甲基四氢甲基蝶呤脱氢酶的基因mtdA (EC 1.5.1.-) ;编码甲基四氢甲基蝶呤环水解酶的基因mch (EC 3.5.4.27) ;编码甲酰基甲烷呋喃四氢甲烷蝶呤N-甲酰基转移酶ftr (EC 2.3.1.101) ;编码甲酰甲烷呋喃脱氢酶的基因fmdA (EC 1.2.7.12) ;编码S-(羟甲基)谷胱甘肽合酶的基因gfa (EC 4.4.1.22) ;编码S-(羟甲基)谷胱甘肽脱氢酶的基因frmA (EC 1.1.1.284) ;编码S-甲酰谷胱甘肽水解酶的基因frmB (EC 3.1.2.12) 。
在CH4氧化过程中,CH4首先被甲烷单加氧酶 (MMO) 转化为甲醇。该过程中MMO可分为可溶性单加氧酶 (sMMO) 和颗粒单加氧酶 (pMMO) ,分别由mmoX (EC 1.14.13.25) 和pmoA (EC 1.14.18.3) 基因编码。生物过滤塔BF_A中pmoA (EC 1.14.18.3) 的相对丰度为0.4%,显著高于BF_no (0.2%) (P≤0.01) ,而2个生物过滤塔中mmoX (EC 1.14.13.25) 的相对丰度没有显著差异 (P>0.05) 。生物过滤塔BF_A中mxaF (EC 1.1.2.7) 的相对丰度为0.6%,极显著高于BF_no (0.2%) (P≤0.001) 。甲醛氧化途径共检测到了3种途径。其中,第一途径中的fae (EC 4.2.1.147) 、mtdA (EC 1.5.1.-) 和fmdA (EC 1.2.7.12) 基因在生物过滤塔BF_A的相对丰度分别为0.9%、1.0%和1.8%,显著高于BF_no (0.4%、0.7%和1.2%) (P≤0.01) 。这表明生物过滤塔BF_A中的CH4代谢活性高于BF_no。
2) 对氮代谢的影响。基于宏基因测序数据,进一步分析了生物过滤塔BF_no和BF_A的氮代谢路径特征,其主要氮代谢路径见图7 (a) ,这说明2个生物过滤塔中具有完整的氮代谢路径。这些基因包括:编码硝酸还原酶的基因narB (EC 1.7.7.2) 、narG (EC 1.7.5.1) 和napA (EC 1.9.6.1) ;编码亚硝酸还原酶的基因nirK (EC 1.7.2.1) ;编码一氧化氮还原酶的基因norB (EC 1.7.2.5) ;编码氧化亚氮还原酶的基因nosZ (EC 1.7.2.4) ;编码同化/异化亚硝酸还原酶的基因nirA (EC 1.7.7.1) 、NIT-6 (EC 1.7.1.4) /nirB (EC 1.7.1.15) 、nrfA (EC 1.7.2.2) ;编码氨单加氧酶的基因amoA (EC 1.14.99.39) ;编码羟胺氧化还原酶的基因hao (EC 1.7.2.6) ;编码固氮酶的基因nifD (EC 1.18.6.1) 。将2个生物过滤塔中的氮代谢关键酶含量进行差异性比较,结果如图7 (b) 所示。在硝化途径中,生物过滤塔BF_A中amoA (EC 1.14.99.39) 和hao (EC 1.7.2.6) 基因的相对丰度分别为0.8%和0.4%,显著高于对照生物过滤塔BF_no (0.4%和0.2%) (P≤0.05) 。这表明生物过滤塔BF_A的硝化过程较活跃。此外,narG (EC 1.7.5.1) 和nirK (EC 1.7.2.1) 基因在2个生物过滤塔的氮转化基因中表达丰度最高,亦证明2个生物过滤塔中均有活跃的反硝化过程。
-
生物过滤技术是净化畜禽养殖场粪尿存储排放的含CH4废气的潜在有效技术,而氮源供应是生物过滤塔高效稳定净化养殖场含CH4废气的关键因素,硝态氮常被用来作为甲烷氧化菌生长代谢过程的营养物质,而铵态氮被认为有可能会对甲烷氧化纯菌的生长产生抑制作用[32],其对以混合菌挂膜启动的生物过滤塔CH4净化性能的影响尚不清晰。本研究结果给出了这一科学问题的答案。铵态氮不会对生物过滤塔的CH4氧化性能产生抑制作用,相反,其还能显著提高生物过滤塔的CH4净化性能,当生物过滤塔的停留时间EBRT缩短至44 min时,以铵态氮供应的生物过滤塔BF_A的CH4去除效率能够稳定在80%以上,而对照生物过滤塔BF_no的CH4去除效率则低不足70%。这说明铵态氮供应能减少除CH4生物过滤塔的占地面积,这具有非常重要的工程应用价值。一直以来,停留时间过长是限制生物过滤法净化CH4的一个关键问题,文献报道中的EBRT可长达1 162 min[33]。缩短停留时间可进一步降低该技术的运行费用,促进其在畜禽养殖业CH4排放控制中的应用和推广。
本研究的宏基因组学分析结果诠释了铵态氮提高生物过滤塔的CH4净化能力的机理。生物过滤塔BF_A中特有的硝化螺旋菌门 (Nitrospirae) 微生物能将铵氮转化为硝态氮[34],从而能解除铵态氮对甲烷氧化菌的抑制,并能提供甲烷氧化菌生长代谢所需要的硝酸盐,从而促进甲烷氧化菌的生长,进而提高生物过滤塔BF_A的CH4氧化能力。变形菌门 (Proteobacteria) 在生物过滤塔BF_A中的相对丰度显著高于对照生物过滤塔BF_no。已知文献均表明,绝大部分甲烷氧化菌都属于变形菌门[35]。由此推断,生物过滤塔BF_A中存在能氧化CH4的大量微生物。此外,基于宏基因组数据绘制CH4代谢通路图,揭示出涉及到CH4代谢的基因pmoA (EC 1.14.18.3) 、mxaF (EC 1.1.2.7)、fae (EC 4.2.1.147)、mtdA (EC 1.5.1.-) 和fmdA (EC 1.2.7.12) 的相对丰度均显著高于对照生物过滤塔BF_no。上述基因组成了一个完整的CH4代谢通路[36],这说明生物过滤塔BF_A中涉及到CH4氧化的各种关键基因都得到了表达,为生物过滤塔BF_A具备较高的CH4氧化能力提供了有力的微生物学证明。
综上所述,以垃圾填埋场覆土作为菌源,可保证了生物过滤塔中的微生物种群非常丰富,而以铵态氮供给生物过滤塔能够实现硝化细菌和反硝化细菌的定向选择。上述微生物与甲烷氧化菌组成的生态系统能保证生物过滤塔较高的CH4氧化能力。本研究结果可为将养殖场高浓度含氮废水应用于生物除CH4过程提供了初步的试验证据,未来应进一步考察实际养殖废水对生物过滤塔CH4氧化性能的影响,为基于生物过滤技术实现养殖场水气污染协同控制提供技术支持。
-
1) 以铵态氮作为氮源的生物过滤塔BF_A的CH4去除性能优于对照生物过滤塔BF_no,当EBRT为44 min时,生物过滤塔BF_A的CH4去除效率仍保持在80%以上,而对照生物过滤塔BF_no的CH4去除效率则不足70%。
2) 宏基因组分析结果表明:2个生物过滤塔的微生物群落结构和物种组成具有显著差异,其中硝化螺旋菌门 (Nitrospirae) 是生物过滤塔BF_A的特有菌门;生物过滤塔BF_A中与CH4代谢和硝化过程有关的基因丰度显著高于对照生物过滤塔BF_no,从微生物学角度佐证了硝化微生物的存在,缓解了铵态氮与CH4竞争MMO活性位点,保证了生物过滤塔较高的CH4氧化效率。
铵态氮对生物过滤塔甲烷净化性能的影响及其微生物学机理
Effects of ammonium nitrogen on methane purification performance of biofilter and its microbiological mechanism
-
摘要: 氮源是生物过滤塔高效稳定净化甲烷 (CH4) 废气的关键因素,然而关于畜禽养殖废水中铵氮是否可以作为除CH4生物过滤塔氮源以及相应的作用机制尚不明晰。启动并成功运行2个生物过滤塔BF_no (对照,无氮源) 和BF_A (铵态氮为氮源) ,比较分析了不同停留时间 (EBRT) 下,2个生物过滤塔的CH4净化性能,并采用宏基因组技术解析了铵态氮影响生物过滤塔CH4净化性能的微生物学机理。结果表明,生物过滤塔BF_A的CH4去除性能优于生物过滤塔BF_no,当EBRT为44 min时,BF_A的CH4去除效率稳定在80%以上,而生物过滤塔BF_no的CH4去除效率不足70%。宏基因组分析结果表明:BF_no和BF_A具有显著不同的微生物群落结构,其中硝化螺旋菌门 (Nitrospirae) 是生物过滤塔BF_A中的特有菌属。生物过滤塔BF_A中与硝化过程相关的amoA和hao基因,以及与CH4氧化相关的fae、mtdA和fmdA基因相对丰度均显著高于BF_no (P≤0.05) ,证明以铵态氮为氮源的生物过滤塔BF_A中不仅具有较高的硝化能力,还具有较高的CH4氧化能力。本研究结果可为生物过滤法净化畜禽养殖含CH4废气和液态养殖粪污的综合利用提供参考。Abstract: Nitrogen is the key factor for the efficient and stable operation of biofilters treating methane (CH4) containing gases. However, it is unclear whether the livestock wastewater with high concentrations of NH4+-N can be used as the nitrogen source of biofilter for CH4 removal and the corresponding mechanism. This study successfully operated two biofilters treating simulated CH4 containing gases, BF_no (control, no nitrogen source) and BF_A (NH4+-N as nitrogen source), and compared CH4 purification performances of the two biofilters under different empty bed residence time (EBRT). The corresponding microbiological mechanism was investigated based on metagenomic analysis. The results showed that BF_A exhibited a better CH4 purification performance in comparison with BF_no. When the EBRT was 44 min, the CH4 removal efficiency of BF_A was stable at more than 80%, while that of BF_no was less than 70%. The two biofliters had significantly different microbial community structures. The species Nitrospirae showed a quite high relative abundance in BF_A. The relative abundance of amoA and hao genes related to nitrification process, and the relative abundance of fae , mtdA and fmdA genes related to CH4 oxidation in BF_A were significantly higher than that in BF_no ( P ≤0.05), which proved that BF_A with NH4+-N as nitrogen source had higher nitrification and CH4 oxidation ability.
-
Key words:
- ammonia nitrogen /
- CH4 oxidation /
- biofiltration /
- metagenome
-

-
-
[1] FERNANDEZ-AMADOR O, OBERDABERNIG D A, TOMBERGER P. Do methane emissions converge? Evidence from global panel data on production-and consumption-based emissions[J]. Empirical Economics, 2022, 63(2): 877-900. doi: 10.1007/s00181-021-02162-9 [2] 汪开英, 李鑫, 陆建定, 等. 碳中和目标下畜牧业低碳发展路径[J]. 农业工程学报, 2022, 38(1): 230-238. [3] MALDANER L, WAGNER-RIDDLE C, VANDERZAAG A C, et al. Methane emissions from storage of digestate at a dairy manure biogas facility[J]. Agricultural and Forest Meteorology, 2018, 258: 96-107. doi: 10.1016/j.agrformet.2017.12.184 [4] 徐文倩, 董红敏, 尚斌, 等. 典型畜禽粪便厌氧发酵产甲烷潜力试验与计算[J]. 农业工程学报, 2021, 37(14): 228-234. [5] DE PUE D, BUYSSE J. Safeguarding natura 2000 habitats from nitrogen deposition by tackling ammonia emissions from livestock facilities[J]. Environmental Science & Policy, 2020, 111: 74-82. [6] RAMIREZ A A, GARCIA-AGUILAR B P, JONES J P, et al. Improvement of methane biofiltration by the addition of non-ionic surfactants to biofilters packed with inert materials[J]. Process Biochemistry, 2012, 47(1): 76-82. doi: 10.1016/j.procbio.2011.10.007 [7] 尚斌, 周谈龙, 董红敏, 等. 生物过滤法去除死猪堆肥排放臭气效果的中试[J]. 农业工程学报, 2017, 33(11): 226-232. doi: 10.11975/j.issn.1002-6819.2017.11.029 [8] SMIRNOVA A V, DUNFIELD P F. Differential transcriptional activation of genes encoding soluble methane monooxygenase in a facultative versus an obligate methanotroph[J]. Microorganisms, 2018, 6(1): 20. doi: 10.3390/microorganisms6010020 [9] GOMEZ-CUERVO S, ALFONSIN C, HERNANDEZ J, et al. Diffuse methane emissions abatement by organic and inorganic packed biofilters: Assessment of operational and environmental indicators[J]. Journal of Cleaner Production, 2017, 143: 1191-1202. doi: 10.1016/j.jclepro.2016.11.185 [10] CHIEMCHAISRI W, CHIEMCHAISRI C, BOONCHAIYUTTASAK J. Utilization of stabilized wastes for reducing methane emission from municipal solid waste disposal[J]. Bioresource Technology, 2013, 141: 199-204. doi: 10.1016/j.biortech.2013.03.035 [11] HUETE A, DE L C D, GOMEZ-BORRAZ T, et al. Control of dissolved CH4 in a municipal UASB reactor effluent by means of a desorption-Biofiltration arrangement[J]. Journal of Environmental Management, 2018, 216: 383-391. [12] HAN J S, MAHANTY B, YOON S U, et al. Activity of a methanotrophic consortium isolated from landfill cover soil: Response to temperature, pH, CO2, and porous adsorbent[J]. Geomicrobiology Journal, 2016, 33(10): 878-885. doi: 10.1080/01490451.2015.1123330 [13] SYED R, SAGGAR S, TATE K, et al. Assessing the performance of floating biofilters for oxidation of methane from dairy effluent ponds[J]. Journal of Environmental Quality, 2017, 46(2): 272-280. doi: 10.2134/jeq2016.08.0310 [14] FERDOWSI M, VEILLETTE M, RAMIREZ A A, et al. Performance evaluation of a methane biofilter under steady state, transient state and starvation conditions[J]. Water Air and Soil Pollution, 2016, 227(6): 168. doi: 10.1007/s11270-016-2838-7 [15] FARROKHZADEH H, HETTIARATCHI J P A, JAYASINGHE P, et al. Aerated biofilters with multiple-level air injection configurations to enhance biological treatment of methane emissions[J]. Bioresource Technology, 2017, 239: 219-225. doi: 10.1016/j.biortech.2017.05.009 [16] LEBRERO R, LOPEZ J C, LEHTINEN I, et al. Exploring the potential of fungi for methane abatement: Performance evaluation of a fungal-bacterial biofilter[J]. Chemosphere, 2016, 144: 97-106. doi: 10.1016/j.chemosphere.2015.08.017 [17] WANG H, QI X, CHEN S, et al. The efficient treatment of breeding wastewater by an electroactive microbial community in microbial fuel cell[J]. Journal of Environmental Chemical Engineering, 2022, 10(2): 107187. doi: 10.1016/j.jece.2022.107187 [18] CHOI H, RYU H W, CHO K S. Biocomplex textile as an alternative daily cover for the simultaneous mitigation of methane and malodorous compounds[J]. Waste Management, 2018, 72: 339-348. doi: 10.1016/j.wasman.2017.11.017 [19] NIKIEMA J, GIRARD M, BRZEZINSKI R, et al. Biofiltration of methane using an inorganic filter bed: Influence of inlet load and nitrogen concentration[J]. Canadian Journal of Civil Engineering, 2009, 36(12): 1903-1910. doi: 10.1139/L09-144 [20] LA H, HETTIARATCHI J P A, ACHARI G, et al. Biofiltration of methane using hybrid mixtures of biochar, lava rock and compost[J]. Environmental Pollution, 2018, 241: 45-54. doi: 10.1016/j.envpol.2018.05.039 [21] HUBER-HUMER M, TINTNER J, BOHM K, et al. Scrutinizing compost properties and their impact on methane oxidation efficiency[J]. Waste Management, 2011, 31(5): 871-883. doi: 10.1016/j.wasman.2010.09.023 [22] NYERGES G, HAN S K, STEIN L Y. Effects of ammonium and nitrite on growth and competitive fitness of cultivated methanotrophic bacteria[J]. Applied and Environmental Microbiology, 2010, 76(16): 5648-5651. doi: 10.1128/AEM.00747-10 [23] WU Y M, YANG J, FAN X L, et al. Elimination of methane in exhaust gas from biogas upgrading process by immobilized methane-oxidizing bacteria[J]. Bioresource Technology, 2018, 260: 432. doi: 10.1016/j.biortech.2018.04.034 [24] CHEN S, ZHOU Y, CHEN Y, et al. Fastp: An ultra-fast all-in-one FASTQ preprocessor[J]. Bioinformatics, 2018, 34(17): 884-890. doi: 10.1093/bioinformatics/bty560 [25] LI D, LIU C M, LUO R, et al. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph[J]. Bioinformatics, 2015, 31(10): 1674-1676. doi: 10.1093/bioinformatics/btv033 [26] NOGUCHI H, PARK J, TAKAGI T. MetaGene: Prokaryotic gene finding from environmental genome shotgun sequences[J]. Nucleic Acids Res, 2006, 34(19): 5623-5630. doi: 10.1093/nar/gkl723 [27] FU L, NIU B, ZHU Z, et al. CD-HIT: accelerated for clustering the next-generation sequencing data[J]. Bioinformatics, 2012, 28(23): 3150-3152. doi: 10.1093/bioinformatics/bts565 [28] LI R, LI Y, KRISTIANSEN K, et al. SOAP: short oligonucleotide alignment program[J]. Bioinformatics, 2008, 24(5): 713-714. doi: 10.1093/bioinformatics/btn025 [29] BUCHFINK B, XIE C, HUSON D H. Fast and sensitive protein alignment using DIAMOND[J]. Nature Methods, 2015, 12(1): 59-60. doi: 10.1038/nmeth.3176 [30] BRANDT E M F, DUARTE F V, VIEIRA J P R, et al. The use of novel packing material for improving methane oxidation in biofilters[J]. Journal of Environmental Management, 2016, 182: 412-420. doi: 10.1016/j.jenvman.2016.07.075 [31] STONE K, HILLIARD M, BADR K, et al. Comparative study of oxygen-limited and methane-limited growth phenotypes of Methylomicrobium buryatense 5GB1[J]. Biochemical Engineering Journal, 2020, 161: 107707. doi: 10.1016/j.bej.2020.107707 [32] LA H, HETTIARATCHI J P A, ACHARI G, et al. Investigation of biologically stable biofilter medium for methane mitigation by methanotrophic bacteria[J]. Journal of Hazardous, Toxic, and Radioactive Waste, 2018, 22(3): 04018013. doi: 10.1061/(ASCE)HZ.2153-5515.0000406 [33] BERGER J, FORNES L V, OTT C, et al. Methane oxidation in a landfill cover with capillary barrier[J]. Waste Management, 2005, 25(4): 369-373. doi: 10.1016/j.wasman.2005.02.005 [34] YANG Z, ZHOU Q, SUN H, et al. Metagenomic analyses of microbial structure and metabolic pathway in solid-phase denitrification systems for advanced nitrogen removal of wastewater treatment plant effluent: A pilot-scale study[J]. Water Research, 2021, 196: 117067. doi: 10.1016/j.watres.2021.117067 [35] LAN H, THI T P, URYNOWICZ M, et al. First investigation of microbial diversity and biogenic methane potential in coal mines located in the Red River Basin, Vietnam[J]. International Journal of Coal Geology, 2021, 234: 103674. doi: 10.1016/j.coal.2020.103674 [36] MA R C, CHU Y X, WANG J, et al. Stable-isotopic and metagenomic analyses reveal metabolic and microbial link of aerobic methane oxidation coupled to denitrification at different O2 levels[J]. Science of the Total Environment, 2021, 764: 142901. doi: 10.1016/j.scitotenv.2020.142901 -



 下载:
下载:







