-
近年来,随着人们对健康水质要求的不断提高,水环境中新兴污染物的环境行为、生态风险及其去除成为研究热点。其中,药品及个人护理品(pharmaceuticals and personal care products,PPCPs)是一类备受关注的新兴污染物[1]。PPCPs主要包括抗生素、激素、镇痛及抗炎药、抗癫痫药物、脂质调节剂、β受体阻滞剂、合成麝香、抗菌剂、防腐剂、防晒霜紫外过滤剂[2]。PPCPs在极低的浓度下即可对生态系统产生不利影响[3-5],由于污水处理厂是PPCPs的主要“源”和“汇”[6],污水中的PPCPs已引起人们的广泛关注[7-8]。
污水生物处理过程,PPCPs通常发生生物及非生物转化,这会导致中间产物生成[9-10]。当前的研究对污水中PPCPs母体化合物关注较多[8, 11],但是对转化产物在污水处理过程的暴露水平及毒性特征却鲜有报道。研究表明,污水中某些PPCPs转化产物较母体的浓度更高[12],如Wang等[13]对污水厂出水中13个药物母体及转化产物进行了定量研究,发现大约40%的产物浓度较母体化合物出水浓度更高。另外,某些产物的毒性较母体化合物更高[14-15],如Fu等[16]指出,经生物转化后,双氯芬酸的甲基化产物较母体化合物的急性毒性提高430倍。因此对污水中PPCPs转化产物的关注尤为必要。
对污水中PPCPs转化产物的识别是探究其在环境水体中暴露水平及毒性特征的基础。产物识别是指对目标PPCPs转化产物进行定性研究,以完全(完整的结构式)或部分(分子量、分子式或不完整的结构式)揭示其结构式的过程[9]。污水生物处理过程,PPCPs转化速度快、转化规律未知。另外污水基质复杂、PPCPs母体及转化产物一般浓度较低[17],这给产物的识别技术提出了较高的要求。目前已有研究对污水中PPCPs转化产物进行识别[18-21],但不同研究间缺乏统一的产物识别流程,其识别的结果难以相互比较,不利于对可能存在的大量未知结构产物的识别,更限制了污水生物处理过程中PPCPs转化产物识别的研究进展。
基于此,本文结合当前污水生物处理系统中PPCPs转化产物的识别研究进展,详细总结了污水中PPCPs转化产物的识别技术,并综述了目前污水中主要PPCPs转化产物的识别进展。在此基础上,对PPCPs转化产物未来的研究方向进行了展望。
-
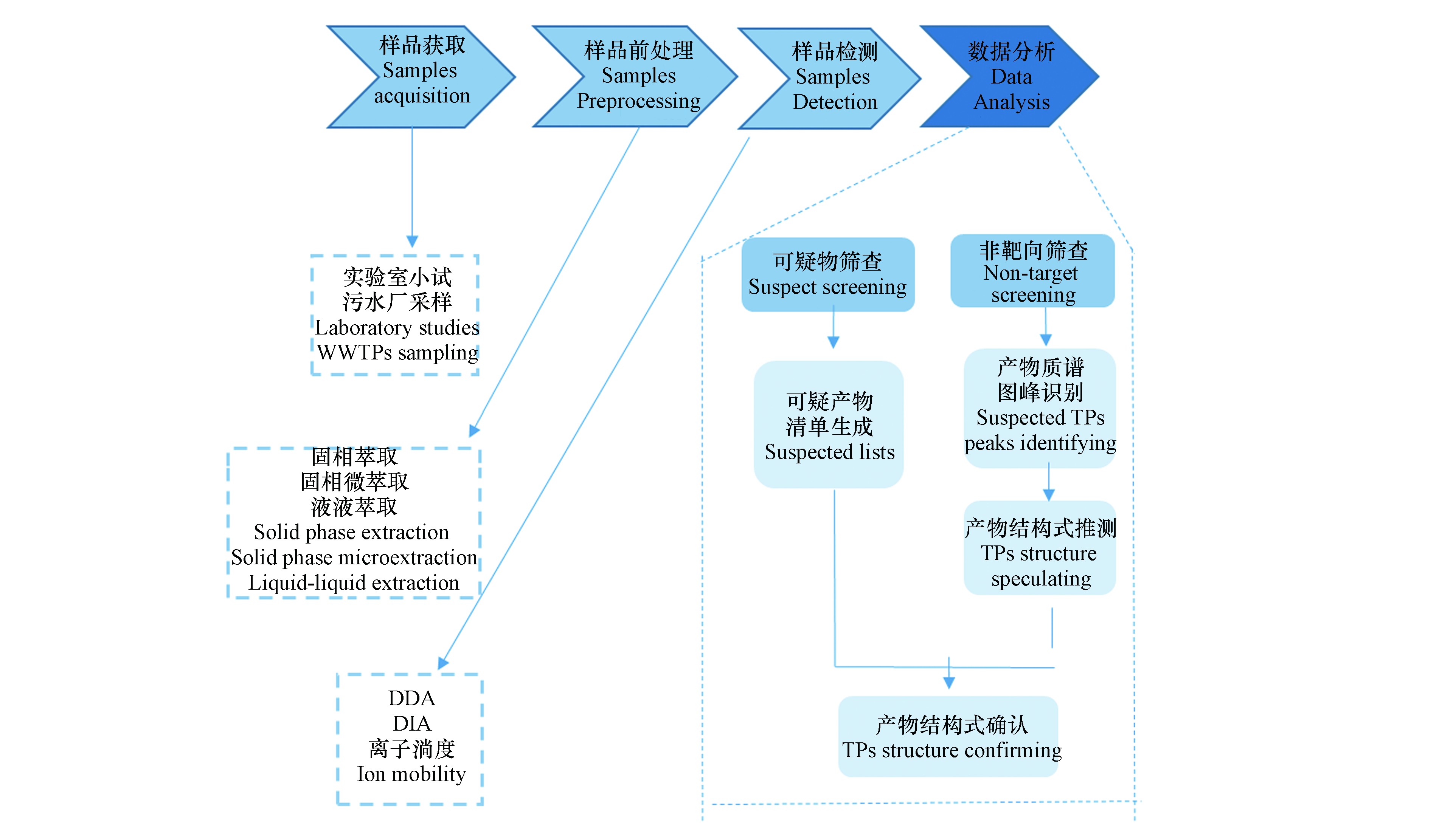
如图1所示,污水中PPCPs转化产物的识别流程一般包括转化产物样品获取[22]、样品前处理、样品上机检测、产物数据分析[23]。目前,以实验室小试实验或者实际污水厂采样获得的识别产物水样较多[24]。获取样品后,由于污水基质复杂,对其进行前处理十分必要。前处理操作一般包括固相萃取、固相微萃取、液液萃取等,其中固相萃取应用最为广泛[25]。对于前处理获取的样品,一般采用液相色谱串联高分辨质谱进行检测,其中数据依赖性采集(data-dependent acquisition,DDA)和数据非依赖性采集(data-independent acquisition,DIA)是液相色谱串联质谱常用的质谱扫描方式[22],另外,离子淌度作为新兴的扫描技术近年来也在环境领域产物同分异构体的鉴别中开始应用[26]。对于获取的产物质谱数据,可以采用可疑物筛查和非靶向筛查策略对产物进行识别[9]。
-
污水中PPCPs转化产物样品获取是对其进行检测及识别的前提。转化产物识别的样品与一般污染物识别或筛查所需的样品要求不同。产物识别的样品需要容易将所识别出产物与目标母体化合物进行关联,以避免所识别出的产物难以归属为哪个母体化合物的难题。通过实验室小试实验(设置空白药物对照组与添加药物的实验组)获取水样是最常见的产物识别样品来源[23],通过对比空白组与实验组中所检出产物的不同即可识别出目标PPCPs的转化产物。另外,实验室小试实验可以获得待研究操作参数条件下PPCPs转化产物样品。通过设置实验室小试实验的操作条件,研究者们已经探究了溶解氧浓度(缺氧、厌氧、好氧)[27]、进水中目标PPCPs浓度[28]、生物处理系统(活性污泥系统、生物膜系统)[29]对PPCPs在污水中转化过程的影响,识别出了不同条件下目标PPCPs的转化产物。然而实验室小试实验在运行规模及进水条件上与实际污水厂均存在差别,因此该条件下获取的样品是否可以真实反映污水中PPCPs的转化还有待商榷[30- 31]。直接采集污水厂水样进行检测识别是另一种产物识别的水样获取方式。然而,由于污水基质过于复杂,多种母体PPCPs共存,这可能会导致在同时存在某些结构类似母体PPCPs的情况下,转化产物来源于哪个母体难以判别。如Chen等[32]在识别经氯消毒后的污水中PPCPs转化产物后,必须通过室内小试实验验证方可确认该产物来源于哪个母体化合物。因此,通过污水厂采样获取样品的方式对PPCPs转化产物的识别研究中,可以确认的转化产物数量一般较少[13, 33]。
结合实验室小试及实际污水厂采样是准确识别污水中PPCPs转化产物较好的样品获取方式。即先通过实验室小试实验尽可能多的识别PPCPs的未知转化产物,再通过对污水厂采样样品中所识别出的产物在污水处理厂的检出情况从而判断实际污水中该转化途径是否发生。通过实验室小试实验获取样品并结合污水厂采样对所识别产物进行验证的方式,德国联邦水文研究所Thomas教授团队已经对污水中双氯芬酸[29]、加巴喷丁[34]、碘普罗胺[35]、甲氧苄氨嘧啶[28]、糖皮质激素[20]、抗病毒药物[36]等一系列PPCPs在污水中的转化产物进行了研究,全面真实的反映了其在污水处理过程中的转化特征。
-
污水水样基质复杂,且PPCPs及其转化产物浓度一般较低,合适的样品前处理可以有效富集目标化合物并除去样品中的部分杂质,有利于进一步的上机检测。转化产物样品前处理的要求与一般的污水中PPCPs靶向筛查或检测不同。靶向筛查中,目标PPCPs性质已知,这使得其选择具有针对性的前处理方式及萃取溶剂或柱型较为明确。然而,由于污水中PPCPs经生物转化生成的产物的具体物理化学性质在识别之前未知,这给合适前处理方法及萃取材料选型带来了困难。当前,常用的样品前处理技术主要包括固相萃取、液液萃取、固相微萃取[37]。固相萃取由于对各种化合物普适性较好且操作流程简单,是目前最常用的前处理方式[25]。固相萃取需要根据目标化合物的物理化学性质(pKa、lgKow等)选择合适的萃取小柱。由于PPCPs一般极性较低,亲水亲油平衡(HLB)固相萃取柱在污水中PPCPs及其转化产物的前处理中使用最为广泛[38]。由于PPCPs转化产物性质未知,且不同产物可能性质各异,一种型号的小柱一般满足不了不同产物的均较好的回收。为了全面回收可能具有不同物理化学性质的转化产物,采用多根不同类型的小柱串联是一种较好的固相萃取方式。Moschet等[39]考察了HLB、弱阴离子交换柱(WAX)、弱阳离子交换柱(WCX)、极性化合物(ENV+)小柱串联对污水中不同形态(阴离子、阳离子、中性)及性质(pKa、lgKow、官能团类型)的新兴污染物的回收效果,发现对于45种目标化合物,91%的化合物回收率在75%到125%之间。Gago-Ferrero等[40]利用聚合物吸附小柱(Strata-X)、阳离子交换聚合物萃取柱(Strata-X-CW)、阴离子交换聚合物萃取柱(Strata-X-AW)、ENV+小柱串联对173种性质不同的化合物进行回收并检测,94%的化合物通过该前处理方式可以被正常检测到。
-
经固相萃取前处理后的转化产物样品浓度一般仍然较低,需要灵敏度较高的检测技术。液相色谱串联高分辨质谱(LC-HRMS/MS)以其较高的灵敏度、精确度被广泛应用于PPCPs转化产物的识别研究中。当前,最常用的LC-HRMS/MS机型包括LC-Q-TOF、LC-Q-Orbitrap及LC-LTQ-Orbitrap[41-42]。
串联质谱通过对目标化合物前驱离子及碎片离子的扫描从而获取其分子量及结构信息。对前驱离子及碎片离子信息同时获取的扫描方式包括DDA和DIA[43]。DDA是指通过对每一时刻全扫数据里相对强度较高的某些前驱离子(一般10个左右)进行打碎从而获取其碎片信息。通过设置动态背景扣除及排除离子对(无需关注的离子对)等参数可以获取更多目标物质的二级质谱碎片信息,从而有利于转化产物结构的解析[44]。目前,DDA采集模式在环境领域化合物的筛查方面使用较多。
然而,由于某些转化产物可能丰度较低,在DDA扫描下难以得到二级碎片信息,这为进一步的结构解析带来困难[45]。DIA扫描可以解决这一问题。DIA是一种对前驱离子根据质量范围分窗格(25 Da)的扫描方式。依赖于TOF质谱较快的扫描速度,从而获取整个质量范围内前驱离子的二级碎片信息[46]。DIA采集技术在蛋白质组学及代谢组学中肽段及代谢物的结构注释方面已经得到广泛的应用。当前,以Swath(sequential window acquisition of all theoretical fragment-ion spectra)为代表的DIA技术已得到较广泛的应用[47- 48]。并且,在第一代固定窗口的基础上,已经开发出根据样品在不同时间内前驱离子数目计算优化而得到的可变窗口[49],这进一步保证了二级谱图的质量,为转化产物的结构解析提供了基础。
PPCPs在污水处理过程中的转化可能会生成同分异构体,色谱对同分异构体的分离能力有限。离子淌度质谱有助于解决产物中同分异构体的识别问题[26]。离子淌度质谱是基于离子淌度分离与质谱联用的新型质谱。在离子漂移管内,离子与缓冲气碰撞时碰撞截面不同,可根据离子大小及形状极性进行分离[17]。目前离子淌度质谱在环境领域应用还不多,但由于其对同分异构体优异的鉴别及在代谢组学代谢物注释方面所表现的优异能力[50-51],必将为环境领域新型化合物的非靶向筛查方面带来新的活力。
-
PPCPs转化产物样品经质谱采集后的数据一般十分复杂,需要合适的谱图解析策略对其数据进行处理从而识别出转化产物的质谱峰并鉴定出产物的结构式。目前,可疑物筛查和非靶向筛查策略是产物识别常用的数据分析技术[9, 52]。
-
可疑物筛查是指在不具备目标PPCPs转化产物标准品的条件下,对已知分子式及结构式的转化产物进行识别确认的过程[9]。可疑物筛查最重要步骤是得到目标PPCPs的转化产物清单。一般可以通过文献检索及软件预测PPCPs可能的转化产物,进而对其谱图提取并识别[13]。
文献调研为PPCPs转化产物研究提供了丰富的可疑产物清单。Wang等[13]基于文献调研生成了农药和医药的转化产物清单,识别出污水中目标药物的56种转化产物。转化产物的预测软件一般基于有机化合物在生物体内的转化规则预测并提供PPCPs转化产物的可疑物清单[53]。有机化合物生物转化产物的预测软件有瑞士水联邦研究所开发的EAWAG BBD/PPS和日本京都大学开发的PathPred预测系统[52, 54],其中EAWAG BBD/PPS在污水生物处理过程中的产物预测使用最为广泛。Huntscha等[55]使用EAWAG BBD/PPS对污水中苯并三唑转化产物进行了预测,并识别出42种转化产物。Beretsou等[53]结合EAWAG BBD/PPS预测可疑产物及非靶向筛查策略,识别出西酞普兰在污水中的14种主要转化产物。但是由于EAWAG BBD/PPS在预测产物时结合了数据库里众多的转化规则,在对PPCPs转化产物进行预测时可能会因“组合爆炸”而生成过多的可疑产物,从而产生假阳性。目前对这一问题还没有很好的解决策略,一般只能通过适当减少预测的层级数目来减少预测产物数目,从而减少不必要的假阳性输出[19]。
-
非靶向筛查是指在对目标PPCPs转化产物不具备任何分子式及结构式的信息下,完全依赖于对实验谱图的解析从而识别其转化产物[9]。转化产物的非靶向筛查中,首先需要在质谱的全扫数据中识别出目标PPCPs的转化产物质谱峰,在此基础上,再根据其二级碎片信息推测确定其结构式[56]。
PPCPs转化产物质谱峰的识别与一般的污水中PPCPs识别的区别在于,所需识别出的质谱峰为目标PPCPs的产物峰,因此,通过实验设计或PPCPs母体及产物间元素、结构的关联性是准确识别出产物峰的关键线索。由于污水样品基质复杂,即使经过固相萃取前处理后,经高分辨质谱得到的全扫峰依旧“杂乱无章”,这使得在众多的质谱峰中(一般大于10000个)寻找产物峰十分困难[9]。目前多结合代谢组学[57]、天然产物识别[58]、新兴污染物识别[57]等领域非靶向筛查相关的知识,通过扣除空白、同位素模式、质量亏损、质谱峰随时间变化趋势、特征碎片、母体与产物结构类似的分子网络等过滤方式对全扫数据中可能的产物峰进行识别[59-61]。
扣除空白是指对实验组的质谱数据进行扣除空白组(不添加目标PPCPs,与实验组其他实验操作条件完全相同)质谱数据处理,以期望扣除那些“杂乱无章”的干扰质谱峰,大大削减全扫数据中质谱峰的数目从而有利于产物峰的识别。Moreira等[62]采用扣除空白的非靶向筛查策略对双氯芬酸在污水中转化产物进行了识别,识别出了12个产物质谱峰并对其结构进行了解析。然而,经空白扣除后,质谱峰一般仍较多,难以确认产物质谱峰。但由于其操作简单,目前扣除空白已成为非靶向筛查数据处理流程中的首要步骤[40]。
同位素模式识别是指目标PPCPs由于其本身所带的具有明显同位素特征的元素(如氯原子)使其产物一级质谱峰成簇出现(同位素峰),通过对同位素峰的识别可以判断出哪个质谱峰为产物峰[32]。一般来说,由于氯及溴原子的不同同位素比例相差较大,可以在质谱谱图上显现,带这两种元素的目标PPCPs的转化产物可以通过同位素模式识别转化产物的质谱峰。Liu等[63]根据此方法识别出了四溴双酚A一系列的转化产物。然而,大多数PPCPs其母体上不带氯及溴原子,同位素模式过滤方式对这些PPCPs产物峰的识别不起作用。
质量亏损过滤原理在于转化产物与母体化合物一般质量亏损值(与最邻近精确质量数的整数值相差的数量)相差不大,因此可以通过设置一段较小的亏损值范围从而过滤出产物的质谱峰[64]。Yu等[65]利用此方法识别出了一系列新的全氟化合物转化产物。
质谱峰丰度随时间变化的识别方法原理在于实验室的小试实验中,实验组中目标PPCPs产物在实验开始前浓度为零,随着实验进行,会出现浓度一直升高或先升高后下降等趋势。这种浓度的变化趋势可以在质谱峰强度上反映。因此可以根据此变化趋势识别出产物的质谱峰。Weizel等[56]通过观察到明显增加或减少趋势的质谱峰且该质谱峰在连续3个实验组样品中均检出,则将其视为产物峰。基于此方式识别出了41种糖皮质激素的转化产物。
特征碎片及母体与产物结构类似的分子网络等过滤方式原理在于具有相似结构式的化合物经质谱碎裂后,会产生相似的二级谱图[66]。因此可以根据类似的二级谱图来识别出结构类似化合物的质谱峰。选取某个具有特征的共性碎片对产物质谱峰识别即为特征碎片过滤方式[59]。选取所有的二级碎片信息并根据余弦相似性得出的相似度构建分子网络即为利用分子网络的过滤方式[67]。Wang等[13]利用特征碎片的过滤方式识别了污水中50种农药及医药的转化产物。而分子网络的过滤方法目前仅在环境领域藻毒素的识别[68]及代谢组学中类似结构代谢物的注释[69]上有应用,在PPCPs转化产物的识别方面还有待探究。
识别了PPCPs转化产物可能的一级质谱峰后,即可确定其分子量并根据仪器的允许测量偏差范围计算出分子式。在此基础上,需要根据其二级谱图进一步确认其结构式。
PPCPs转化产物结构式的推测有两种方式。一是根据丢失或增加的分子量推测参与转化的官能团,另外,有机化合物在微生物体系下的转化规则也有利于上述产物结构式的推测[70]。二是根据识别出的产物分子量在化合物数据库中搜索可能的结构式,从而为转化产物的结构解析提供线索[71]。目前化合物数据库中收录的已知化合物信息丰富,ChemSpider中收录了6700万化合物的结构信息,PubChem数据库中目前收录了10300万化合物的结构信息[72],这些化合物中可能含有目标PPCPs转化产物的结构信息。通过与化合物数据库中化合物的搜索可以极大地减小产物结构式推测的难度(对于化合物数据库中存在的转化产物)。
转化产物结构式的确认主要依赖于对二级谱图的解析。与标准品的二级谱图匹配是最可信的结构式确认方式[20]。然而,大多数转化产物无法购买标准品。对于这些转化产物,可以在文献及谱图数据库中检索可能存在的二级谱图信息。目前常用的谱图数据库包括mzCloud、MSBank[73]及METLIN[74],这些谱图数据库中包含了各类化合物的质谱谱图数据,为PPCPs转化产物的结构式确认提供了支撑信息。对于无标准品且数据库中检索不到的转化产物,可以通过谱图预测软件,如根据键能碎裂的MetFrag[75]和根据断裂规则断裂的Mass Frontier[76]进行二级谱图预测,给产物结构确定提供支撑。
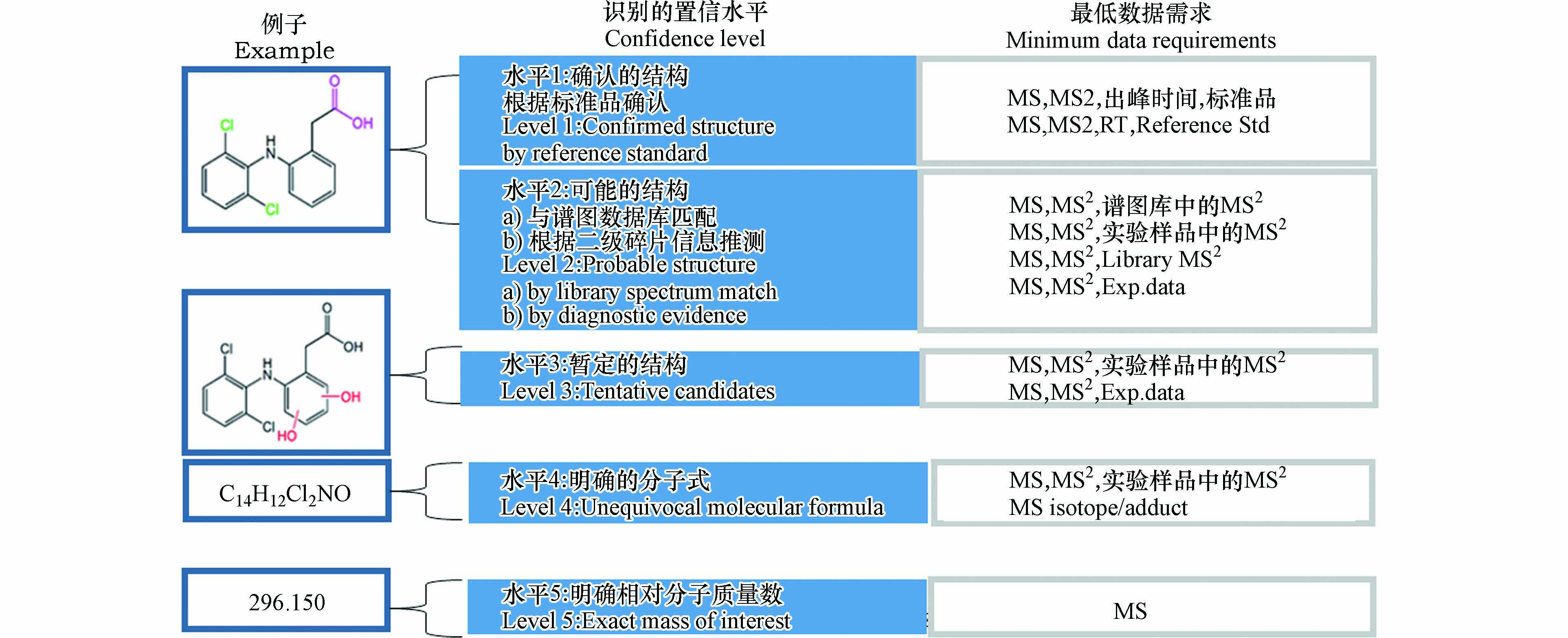
所有PPCPs转化产物经上述流程确认结构后,需要根据图2[77]中的规则进行可信度等级打分。目前该可信度打分标准已成为产物鉴定领域国际公认的评价指标。
-
产物识别流程上,多数研究采用了实验室小试-污水厂采样获取样品、固相萃取前处理、DDA质谱数据采集、产物的可疑物/非靶向筛查方式。但识别技术上,目前多数研究还是使用可疑物筛查技术进行识别,少数采用非靶向筛查的研究仅仅根据质谱峰的相对强度识别产物峰[78],这可能是多数研究中目标抗生素转化产物识别数量较少的原因。对于缺失标准品产物结构式的确认,利用谱图数据库及谱图预测软件预测谱图进行结构确认的方式使用较少。转化反应方面,目前大多数识别出的为Ι型反应的产物,为羟基化、氧化、甲基化、硫化、脱氢、水解等反应产物。下面对不同类型PPCPs转化产物的识别流程及主要识别出的反应分别阐述。
-
抗生素是使用量最大的PPCPs,其化合物自身的负面生态效应及由此引发的抗性基因传播问题引发了人们对其广泛关注[79],目前不同抗生素在污水中的转化产物已被研究(表1)。
Jewell等[28]通过实验室小试实验和污水厂采样获取样品、ENVI-Carb小柱前处理、DDA扫描和可疑物筛查策略识别出6个甲氧苄氨嘧啶的转化产物。其中,羟基化和去甲基是主要的转化反应。另外发现添加高浓度的甲氧苄氨嘧啶(500 μg·L−1)无法真实反映环境条件下痕量有机污染物的转化特征(仅识别出2个产物),且仅发生羟基化反应。Deng等[78]通过实验室小试、非靶向筛查策略识别出12个磺胺嘧啶的转化产物,其中2-氨基-4-羟基嘧啶是主要的转化产物。通过对这些产物转化途径的分析,发现磺胺嘧啶主要通过3条转化途径生成这些产物。其中羟基化和开环是主要的反应步骤。Ooi等[80]通过实验室小试及污水厂采样获得了克林霉素的转化产物样品,经离心过滤处理及质谱检测,识别出3个转化产物,克林霉素主要发生硫化、去甲基、氧化反应,其中硫化的克林霉素是主要转化产物,经生物膜反应器处理1d后,其浓度高达进水中克林霉素的10%。Achermann等[81]通过实验室小试和污水厂采样获得了磺胺甲恶唑转化产物的样品,通过HLB、WAX、WCX、ENV+串联前处理方式、DDA扫描和非靶向筛查策略识别出磺胺甲恶唑的11个转化产物,羟基化、氧化、去碳基、螯合(加蝶呤)是主要的反应类型。值得注意的是,生成蝶呤的螯合反应最近被鉴定为磺胺类抗生素的主要转化方式,其蝶呤螯合物依旧保持抗生素的活性,对水环境造成的持久负面效应不容忽视[82]。Sun等[95]采用实验室小试实验获取了磺胺甲恶唑在硝化污泥中的样品,通过HLB固相萃取前处理,DDA扫描的方式识别出3个转化产物,其发生的反应分别是硝化、脱氨基和甲基化。另外发现这3种产物的生成与游离亚硝酸的浓度正相关,当游离亚硝酸浓度降低时,这些产物可以进一步逆转化成磺胺甲恶唑母体。
-
激素类PPCPs包括天然及人工合成的对人类内分泌系统产生干扰作用的化合物,由于其具有改变哺乳动物性别性状的潜力而引起人们的关注。各种激素类药物在污水中的转化已被广泛报道(表1)。
Yu等[83]通过实验室小试实验获取孕酮在传统好氧活性污泥和添加了功能微生物的好氧活性污泥中转化产物的样品,固相萃取前处理、可疑物筛查等方式识别出孕酮的11(功能微生物强化的好氧活性污泥中)及12个转化产物(传统好氧活性污泥中),发现羟基化、氧化、开环是其主要的反应类型。Liu等[84]采用实验室小试、HLB固相萃取前处理方式识别了孕酮和去氢孕酮在养猪废水中的转化,分别识别出8个孕酮及6个去氢孕酮的转化产物。其中,羟基化、氧化、还原、侧链断裂是孕酮和去氢孕酮的主要反应类型。同时作者指出,虽然孕酮和去氢孕酮在养猪废水处理中得到了有效的去除,但并非完全矿化,其产物的风险仍值得关注。Weizel等[56]使用实验室小试及污水厂采样获取样品,采用DDA扫描和非靶向筛查策略识别了13种糖皮质激素的41种转化产物。其中,12种可以在污水厂出水中检出,并且8种转化产物在之前的污水厂调研中未见报道。羟基化、甾体C17侧链转化、脂水解、氧化羟基化是主要的反应类型。这些产物可能是污水厂出水中内分泌干扰活性的贡献者。Weizel 等[85]通过实验室小试实验及污水厂采样分析相结合的方式制备了孕激素转化产物水样,采用固相萃取前处理、DDA扫描及非靶向筛查的策略识别出44种羟基孕酮衍生物的转化产物,其中羟基化、环A脱氢反应是主要的反应类型。值得注意的是,环丙氯地孕在污水生物处理中几乎完全转化为3α-羟基环丙氯地孕,这种产物被报道具有遗传毒性。因此,仅关注污水中激素类母体化合物是不够的,需要对其产物的生态风险进行进一步的甄别和削减。
-
非甾体抗炎药是一类消炎镇痛的药物,广泛用于头痛、风湿、骨质疏松等症状的治疗[86]。不同非甾体抗炎药在污水中的转化研究如表1所示。
Wu等[87]通过实验室小试实验获取双氯芬酸在富集硝化污泥、异氧污泥、传统活性污泥中转化产物样品,采用固相萃取前处理、可疑物筛查的策略分别识别出9个(富集硝化污泥系统)、12个(异氧污泥系统)和14个转化产物(传统活性污泥系统)。其中羟基化、脱羧、硫化、氧化是其主要的转化反应。值得注意的是,富集的硝化污泥中,双氯芬酸的羟基化速度更快,但在富集硝化污泥中生成的双氯芬酸苯甲酸这个产物难以进一步转化,只有在异氧微生物的参与下才能进一步转化。因此作者提出污水生物处理过程,硝化微生物和异养微生物可以协同合作转化双氯芬酸。Lu等[88]通过实验室小试实验获取萘普生和布洛芬转化产物样品,识别出7个萘普生和7个布洛芬的转化产物。发现脱羰基、螯合(络氨酸)、羟基化、开环是萘普生的主要转化反应,其中羟基化的产物和羧酸化的产物是布洛芬的主要中间产物。对于萘普生的转化,羟基化、脱羰基、氧化反应参与了布洛芬产物的形成。Bouju等[89]通过添加C14标记的的双氯芬酸探究了其在实验室小试实验污水生物处理体系下的转化,通过固相萃取前处理、DDA扫描及可疑物筛查,识别出5个转化产物,并发现双氯芬酸的羟基化是其转化的关键步骤。Jakimska等[90]采用实验室小试实验和污水厂采样制备了布洛芬和酮洛芬转化产物的样品,利用固相萃取前处理和DDA扫描的方式对产物进行识别,分别识别出13种布洛芬及7种酮洛芬的转化产物。布洛芬主要发生脱羧和羟基化反应,酮洛芬主要发生的是羟基化反应。
-
精神类药物是指用于治疗人体精神方面疾病的药物,如抗癫痫类药物卡马西平、加巴喷丁等[91]。Beretsou等[53]通过实验室小试实验及污水厂采样获取了西酞普兰的转化产物样品,利用DDA质谱扫描方式和可疑物筛查、非靶向筛查方式识别出西酞普兰14个转化产物,其中11种转化产物是新识别出的产物,这些产物中,有5种可以在污水厂出水中检出。氧化、水解、羟基化、去甲基是主要的转化反应。通过污水厂中产物浓度的半定量数据,作者考察了西酞普兰及其产物的毒性,发现其单一的母体或产物其风险商均小于1,但是作者也提到,其产物和母体共存时的联合暴露风险依然不容忽视。Gornik等[24]同样采用实验室小试和污水厂采样获得舍曲林转化产物的样品,用可疑物筛查和非靶向筛查结合的方式识别出舍曲林的10个转化产物,其中8种转化产物可以在污水厂的进出水中检出。羟基化、氧化、螯合是主要的转化反应。Henning等[34]通过实验室小试试验获得了加巴喷丁转化产物的样品,采用固相萃取前处理和DDA扫描的方式识别出8种转化产物,羟基化、氧化、水解是其主要的转化反应。另外,作者发现酰胺化的加巴喷丁是主要的转化产物,在出水中占初始加巴喷丁浓度的18%,并且这种产物难以进一步转化,会在出水中累积。对污水厂出水中加巴喷丁和酰胺化的加巴喷丁定量研究发现,其浓度分别为0.64 mg·L−1和 0.07 mg·L−1。因此,对出水中加巴喷丁产物不容忽视。Konig等[92]通过实验室小试实验获得了卡马西平在污水中转化产物的样品,识别出9种转化产物,加氢、甲基化是其主要的反应类型。其中二氢-卡马西平是主要的转化产物。由于卡马西平较为稳定,还原脱氢是其主要的反应类型,因此作者建议厌氧环境有利于卡马西平的生物转化。Funke等[36]采用实验室小试实验和污水厂调研获取了阿巴卡韦、恩曲他滨、更昔洛韦、拉米夫定和齐多夫定五种精神类药物转化产物的样品,分别识别出这些精神类药物的4种(阿巴卡韦)、3种(恩曲他滨)、4种(更昔洛韦)、3种(拉米夫定)和3种(齐多夫定)转化产物,并发现羧基化反应是这五种精神类药物共有的转化反应。另外,作者对污水厂出水中这些产物的检出浓度表明,其产物在污水中的不容忽视。
-
其他类药物包括脂质调节剂、抗菌剂等。转化产物的识别流程上,最新的产物识别技术(非靶向筛查、化合物数据库搜索、谱库匹配、谱图预测软件)使用较少。转化反应途径上,各类药物结构差别较大,转化反应各异。如Helbling等[52]通过实验室小试实验获得了苯扎贝特和缬沙坦的水样,采用DDA扫描及可疑物筛查、非靶向筛查策略分别识别出苯扎贝特的5个产物和缬沙坦的3个产物,发现加氢、羟基化是苯扎贝特的主要转化反应,脱烷基、水解是缬沙坦的主要转化反应。Henning等[93]通过实验室小试和污水厂采样获得了西他列汀和非索非那定转化产物样品,采用DDA扫描及非靶向筛查策略分别识别出西他列汀9种产物和非索非那定的13种转化产物。并发现水解、螯合是西他列汀的主要转化反应,氧化、羟基化是非索非那定的主要转化反应。Ding等[97]通过实验室小试实验识别了三氯生的转化,发现3个转化产物,主要发生脱氯、羟基化、螯合反应。Armstrong等[98]通过实验室小试及HLB小柱固相萃取的方式并采用DDA采集模式识别了三氯生和三氯卡班在污水生物处理过程中的转化,识别出3种三氯生的转化产物和4种三氯卡班的转化产物,其中三氯生主要发生甲基化反应,三氯卡班主要发生脱氯的反应。
-
本文详细介绍了污水中PPCPs转化产物的识别流程,并对目前污水中不同PPCPs转化产物识别的方法及反应类型进行了概述。基于目前产物识别方面存在的问题,提出以下展望:
(1) 目前PPCPs转化产物识别中使用非靶向筛查策略并结合谱库及谱图预测软件搜索匹配等操作的研究还较少,需要结合这些最新的技术对PPCPs在污水中的转化产物进行全面准确的识别;
(2) PPCPs转化产物在污水中的污染水平研究较少。需要建立PPCPs转化产物的化合物结构及质谱数据谱,结合谱库对污水中PPCPs转化产物进行高通量筛查;
(3) PPCPs转化产物的毒性研究较少,缺乏标准品给产物毒性研究带来巨大困难。可以结合计算毒理学、基于机器学习的毒性预测等方法评估污水中PPCPs转化产物的生态风险。
污水生物处理过程药品及个人护理品转化产物识别研究进展
Identification of transformation products of PPCPs during wastewater biological treatment processes
-
摘要: 药品及个人护理品(pharmaceuticals and personal care products,PPCPs)在污水生物处理过程会发生生物及化学转化从而生成转化产物,某些转化产物较母体化合物毒性更高。目前对PPCPs转化产物关注不够,转化产物识别方法不明确。本文介绍了污水中PPCPs转化产物的识别流程,系统阐述了样品获取、样品前处理、基于液相色谱串联高分辨质谱的样品检测及质谱数据解析等技术,提出了适合污水中PPCPs转化产物的识别技术。在此基础上,分别阐述了抗生素类、激素类、非甾体抗炎药类、精神类药物及其他PPCPs在污水中的产物识别技术及所识别出的产物及转化反应。本综述为全面识别污水中PPCPs转化产物并采取有效的控制策略提供了参考方法。Abstract: Pharmaceuticals and personal care products (PPCPs) are subject to biotic and abiotic transformation in wastewater, which can cause the formation of transformation products (TPs). Some TPs of PPCPs are more toxic than parent compounds. However, the attention paid on TPs is insufficient and the methods on identifying TPs of PPCPs in wastewater are scare. In this study, the workflows of identification of TPs formed from PPCPs during wastewater biological treatment processes were introduced. The details on sample acquisition, sample preparation, detecting by liquid chromatography-high resolution mass spectrometry and data analysis were elaborated. Based on the procedure of identification of TPs mentioned above, the details of identifying TPs from various PPCPs were compared and the proper methods were put forward to fully identifying TPs of PPCPs in wastewater. Accordingly, the identified TPs and involved transformation reactions of various PPCPs such as antibiotic, hormones, non-steroidal anti-inflammatory drugs, psychotropic pharmaceuticals and others were stated in detail. Present review provides a comprehensive picture of TPs from various PPCPs in WWTPs.
-
Key words:
- PPCPs /
- transformation products /
- identification /
- sewage /
- biological treatment
-

-
表 1 不同类型PPCPs在污水生物处理系统中的转化产物识别
Table 1. Identification of transformation products of various PPCPs during wastewater biological treatment
药品及
个人护理品
PPCPs样品获取方式
Methods of
sample acquisition前处理方法
Methods of
sample pretreatment数据采集技术
Techniques of
data acquisition识别技术
Techniques of
TPs identification产物数目
The numbers
of TPs涉及到的反应类型
The involved
reaction types参考
文献
References抗生素 甲氧苄氨嘧啶 实验室小试+
污水厂采样固相萃取
ENVI-Carb小柱DDA 可疑物筛查 6 去甲基化、羟基化、氧化、开环 [28] 磺胺甲恶唑 实验室小试+
污水厂采样固相萃取
HLB、WAX、WCX、ENV+DDA 可疑物筛查 11 羟基化、氧化、去碳基、螯合(加蝶呤) [81] 磺胺嘧啶 实验室小试+
污水厂采样固相萃取
HLB、WAX、WCX、ENV+DDA 可疑物筛查 8 羟基化、氧化、去碳基、螯合(加蝶呤) [81] 磺胺嘧啶 实验室小试 —1 先一级再二级扫描 非靶向筛查 12 羟基化、开环 [78] 甲氧苄氨嘧啶 实验室小试+
污水厂采样固相萃取
ENVI-Carb小柱— 非靶向筛查 6 去甲基化、羟基化、开环、氧化 [28] 克林霉素 实验室小试+
污水厂采样离心、过膜 — — 3 硫化 [80] 磺胺甲恶唑 实验室小试 — — — 4 羟基化、开环 [94] 磺胺甲恶唑 实验室小试 固相萃取HLB DDA — 3 脱氨基反应、羟基化、硝化 [95] 激素类 孕酮 实验室小试 固相萃取HLB 先一级再二级 可疑物筛查 12 羟基化、氧化、开环 [83] 睾酮 实验室小试 固相萃取HLB 先一级再二级 可疑物筛查 15 羟基化、氧化、还原 [27] 孕酮 实验室小试 固相萃取HLB — — 8 羟基化、氧化、还原 [84] 孕激素 实验室小试+
污水厂采样固相萃取HLB DDA 非靶向筛查 44 羟基化、环A脱氢 [85] 去氢孕酮 实验室小试 固相萃取HLB — — 5 羟基化、氧化、加氢 [84] 甲基炔诺酮 实验室小试 固相萃取HLB — — 2 加氢、脱氢 [96] 倍他米松 实验室小试+
污水厂采样— DDA 非靶向筛查 6 脱氢、氧化、硫化、羟基化 [56] 非甾体抗炎药 双氯芬酸 实验室小试+
污水厂采样过膜 DDA 可疑物筛查、
非靶向筛查20 羟基化、去羰基、氧化、酰胺化、开环、还原脱氯 [29] 萘普生 实验室小试 — — — 7 脱羰基、螯合(络氨酸)、羟基化、开环 [88] 布洛芬 实验室小试 — — — 7 羟基化、脱羰基、氧化 [88] 双氯芬酸 实验室小试+
污水厂采样— DDA 可疑物筛查 4 羟基化、脱氢 [89] 双氯芬酸 实验室小试 固相萃取HLB 先一级再二级扫描 可疑物筛查 14 羟基化、酰胺化、硫化、甲基化、氧化 [87] 精神类药物 舍曲林 实验室小试+
污水厂采样固相萃取HLB DDA 非靶向筛查 10 羟基化、氧化、螯合 [24] 加巴喷丁 实验室小试 — DDA — 8 羟基化、氧化、水解 [34] 卡马西平 实验室小试 — — — 9 加氢、甲基化 [92] 西酞普兰 实验室小试+
污水厂采样— DDA 可疑物筛查、
非靶向筛查14 氧化、水解、
羟基化、去甲基[53] 阿巴卡韦 实验室小试+
污水厂采样过膜 — — 4 羧基化、羟基化、去环丙醇 [36] 恩曲他滨 实验室小试+
污水厂采样过膜 — — 3 羧基化、氧化 [36] 更昔洛韦 实验室小试+
污水厂采样过膜 — — 4 羧基化 [36] 拉米夫定 实验室小试+
污水厂采样过膜 — — 3 羧基化、氧化 [36] 齐多夫定 实验室小试+
污水厂采样过膜 — — 3 羧基化、羟基化 [36] 其他 苯扎贝特 实验室小试 — DDA 可疑物筛查、
非靶向筛查5 加氢、羟基化 [52] 缬沙坦 实验室小试 — DDA 可疑物筛查、
非靶向筛查3 脱烷基、水解 [52] 西他列汀 实验室小试+
污水厂采样— DDA 非靶向筛查 9 水解、螯合 [93] 非索非那定 实验室小试+
污水厂采样— DDA 非靶向筛查 13 氧化、羟基化 [93] 三氯生 实验室小试 — DDA — 7 脱氯、羟基化、螯合 [90] 三氯生 实验室小试 固相萃取HLB DDA — 3 甲基化 [97] 三氯卡班 实验室小试 固相萃取HLB DDA — 4 脱氯 [98] 注:1表示文献中无相应值. Note: 1 refers to the information not specified in the reference. -
[1] PETRIE B, BARDEN R, KASPRZYK-HORDERN B. A review on emerging contaminants in wastewaters and the environment: current knowledge, understudied areas and recommendations for future monitoring [J]. Water Research, 2015, 72: 3-27. doi: 10.1016/j.watres.2014.08.053 [2] LIU L, WONG M H. Pharmaceuticals and personal care products (PPCPs): a review on environmental contamination in China [J]. Environment International, 2013, 59: 208-224. doi: 10.1016/j.envint.2013.06.012 [3] OASKS J L, GILBERT M, VIRANI M Z, et al. Diclofenac residues as the cause of vulture population decline in Pakistan [J]. Nature, 2004, 427(6975): 630-633. doi: 10.1038/nature02317 [4] ZHOU S, DI PAOLO C, WU X, et al. Optimization of screening-level risk assessment and priority selection of emerging pollutants - The case of pharmaceuticals in European surface waters [J]. Environment International, 2019, 128: 1-10. doi: 10.1016/j.envint.2019.04.034 [5] DUSZA H M, JANSSEN E, KANDA R, et al. Method development for effect-directed analysis of endocrine dsrupting compounds in human amniotic fluid [J]. Environmental Science & Technology, 2019, 53,(24): 14649-14659. [6] YU Y, HAN P, ZHOU L, et al. Ammonia monooxygenase-mediated cometabolic biotransformation and hydroxylamine-mediated abiotic transformation of micropollutants in an AOB/NOB co-culture [J]. Environmental Science & Technology, 2018, 52(16): 9196-9205. [7] BEN W, ZHU B, YUAN X, et al. Occurrence, removal and risk of organic micropollutants in wastewater treatment plants across China: Comparison of wastewater treatment processes [J]. Water Research, 2018, 130: 38-46. doi: 10.1016/j.watres.2017.11.057 [8] ZHANG Y, WANG B, CAGNETTA G, et al. Typical pharmaceuticals in major WWTPs in Beijing, China: Occurrence, load pattern and calculation reliability [J]. Water Research, 2018, 140: 291-300. doi: 10.1016/j.watres.2018.04.056 [9] BLETSOU A A, JEON J, HOLLENDER J, et al. Targeted and non-targeted liquid chromatography-mass spectrometric workflows for identification of transformation products of emerging pollutants in the aquatic environment [J]. TrAC Trends in Analytical Chemistry, 2015, 66: 32-44. doi: 10.1016/j.trac.2014.11.009 [10] HERNANDEZ F, BAKKER J, BIJLSMA L, et al. The role of analytical chemistry in exposure science: Focus on the aquatic environment [J]. Chemosphere, 2019, 222: 564-583. doi: 10.1016/j.chemosphere.2019.01.118 [11] TRAN N H, CHEN H, REINHARD M, et al. Occurrence and removal of multiple classes of antibiotics and antimicrobial agents in biological wastewater treatment processes [J]. Water Research, 2016, 104: 461-472. doi: 10.1016/j.watres.2016.08.040 [12] EGGEN R I, HOLLENDER J, JOSS A, et al. Reducing the discharge of micropollutants in the aquatic environment: the benefits of upgrading wastewater treatment plants [J]. Environmental Science & Technology, 2014, 48,(14): 7683-7689. [13] WANG X, YU N, YANG J, et al. Suspect and non-target screening of pesticides and pharmaceuticals transformation products in wastewater using QTOF-MS [J]. Environment International, 2020, 137: 105599. doi: 10.1016/j.envint.2020.105599 [14] JAEN-GIL A, CASTELLET-ROVIRA F, LLORCA M, et al. Fungal treatment of metoprolol and its recalcitrant metabolite metoprolol acid in hospital wastewater: Biotransformation, sorption and ecotoxicological impact [J]. Water Research, 2019, 152: 171-180. doi: 10.1016/j.watres.2018.12.054 [15] CHEN K, TIAN F, WU C, et al. Degradation products and pathway of ethiprole in water and soil [J]. Water Research, 2019, 161: 531-539. doi: 10.1016/j.watres.2019.06.004 [16] FU Q, FEDRIZZI D, KOSFELD V, et al. Biotransformation changes bioaccumulation and toxicity of diclofenac in aquatic organisms [J]. Environmental Science & Technology, 2020, 54: 4400-4408. [17] RICHARDSON S D, KIMURA S Y, Water analysis: Emerging contaminants and current issues [J]. Analytical Chemistry, 2020, 92, (1) : 473-505. [18] WOLFENDER J L, NUZILLARD J M, VAN DER HOOFT J J J, et al. Accelerating metabolite identification in natural product research: toward an ideal combination of LC-HRMS/MS and NMR profiling, in silico databases and chemometrics [J]. Analytical Chemistry, 2018, 91,(1): 704-742. [19] GULDE R, MEIER U, SCHYMANSKI E L, et al. Systematic exploration of biotransformation reactions of amine-containing micropollutants in activated sludge [J]. Environmental Science & Technology, 2016, 50,(6): 2908-2920. [20] CASTRONOVO S, WICK A, SCHEURER M, et al. Biodegradation of the artificial sweetener acesulfame in biological wastewater treatment and sandfilters [J]. Water Research, 2016, 110: 342-353. [21] KHALEEL N D H, MAHMOUD W M M, OLSSON O, et al. Studying the fate of the drug Chlorprothixene and its photo transformation products in the aquatic environment: Identification, assessment and priority setting by application of a combination of experiments and various in silico assessments [J]. Water Research, 2019, 149: 467-476. doi: 10.1016/j.watres.2018.10.075 [22] KLONT F, JAHN S, GRIVET C, et al. SWATH data independent acquisition mass spectrometry for screening of xenobiotics in biological fluids: Opportunities and challenges for data processing [J]. Talanta, 2020, 211: 120747. doi: 10.1016/j.talanta.2020.120747 [23] PICO Y, BARCELO D. Transformation products of emerging contaminants in the environment and high-resolution mass spectrometry: a new horizon [J]. Analytical and Bioanalytical Chemistry, 2015, 407(2): 6257-6273. [24] GORNIK T, KOVACIC A, HEATH E, et al. Biotransformation study of antidepressant sertraline and its removal during biological wastewater treatment [J]. Water Research, 2020, 181: 115864. doi: 10.1016/j.watres.2020.115864 [25] RATERINK R J, LINDENBURG P W, VREENKEN R J, et al. Recent developments in sample-pretreatment techniques for mass spectrometry-based metabolomics [J]. TrAC Trends in Analytical Chemistry, 2014, 61: 157-167. doi: 10.1016/j.trac.2014.06.003 [26] WU Q, WANG J Y, HAN D Q, et al. Recent advances in differentiation of isomers by ion mobility mass spectrometry [J]. TrAC Trends in Analytical Chemistry, 2020, 124: 115801. doi: 10.1016/j.trac.2019.115801 [27] YU Q, GENG J, REN H, Occurrence and fate of androgens in municipal wastewater treatment plants in China [J]. Chemosphere, 2019: 124371. [28] JEWELL K S, CASTRONOVO S, WICK A, et al. New insights into the transformation of trimethoprim during biological wastewater treatment [J]. Water Research, 2016, 88: 550-557. doi: 10.1016/j.watres.2015.10.026 [29] JEWELL K S, FALAS P, WICK A, et al. Transformation of diclofenac in hybrid biofilm-activated sludge processes [J]. Water Research, 2016, 105: 559-567. doi: 10.1016/j.watres.2016.08.002 [30] ERICSON J F. Evaluation of the OECD 314B Activated Sludge Die-Away Test for Assessing the Biodegradation of Pharmaceuticals [J]. Environmental Science & Technology, 2010, 44(1): 375-381. [31] MARTIN T J, SNAPE J R, BARTRANM A, et al. Environmentally Relevant inoculum concentrations improve the reliability of persistent assessments in biodegradation screening tests [J]. Environmental Science & Technology, 2017, 51,(5): 3065-3073. [32] CHEN W L, CHENG J Y, LIN X Q. Systematic screening and identification of the chlorinated transformation products of aromatic pharmaceuticals and personal care products using high-resolution mass spectrometry [J]. Science of the Total Environment, 2018, 637-638: 253-263. [33] FREELING F, ALYGIZAKIS N A, VON DER OHE P C, et al. Occurrence and potential environmental risk of surfactants and their transformation products discharged by wastewater treatment plants [J]. Science of the Total Environment, 2019, 681: 475-487. doi: 10.1016/j.scitotenv.2019.04.445 [34] HENNING N, KUNKEL U, WICK A, et al. Biotransformation of gabapentin in surface water matrices under different redox conditions and the occurrence of one major TP in the aquatic environment [J]. Water Research, 2018, 137: 290-300. doi: 10.1016/j.watres.2018.01.027 [35] REDEKER M, WICK A, MEERMANN B, et al. Anaerobic transformation of the iodinated X-ray contrast medium iopromide, its aerobic transformation products, and transfer to further iodinated X-ray contrast media [J]. Environmental Science & Technology, 2018, 52,(15): 8309-8320. [36] FUNKE J, PRASSE C, TERNES T A. Identification of transformation products of antiviral drugs formed during biological wastewater treatment and their occurrence in the urban water cycle [J]. Water Research, 2016, 98: 75-83. [37] OETJEN K, GIDDINGS C G S, MCLAUGHLIN M, et al. Emerging analytical methods for the characterization and quantification of organic contaminants in flowback and produced water [J]. Trends in Environmental Analytical Chemistry, 2017, 15: 12-23. doi: 10.1016/j.teac.2017.07.002 [38] ANDRADE-EIROA A, CANLE M, LEROY-CANCELLIIERRI V, et al. Solid-phase extraction of organic compounds: A critical review (Part I) [J]. TrAC Trends in Analytical Chemistry, 2016, 80: 641-654. doi: 10.1016/j.trac.2015.08.015 [39] MOSCHET C, PIAZZOLI A, SINGER H, et al. Alleviating the reference standard dilemma using a systematic exact mass suspect screening approach with liquid chromatography-high resolution mass spectrometry [J]. Analytical Chemistry, 2013, 85,(21): 10312-10320. doi: 10.1021/ac4021598 [40] GAGO-FERRERO P, SCHYMANSHI E L, BLETSOU A A, et al. Extended suspect and non-target strategies to characterize emerging polar organic contaminants in raw wastewater with LC-HRMS/MS [J]. Environmental Science & Technology, 2015, 49,(20): 12333-12341. [41] PRASSE C, TERNES T A. Application of orbitrap mass spectrometry for the identification of transformation products of trace organic contaminants formed in the environment [J]. Comprehensive Analytical Chemistry, 2016, 71: 282. [42] BOIX C, IBANEZ M, BAGNATI R, et al. High resolution mass spectrometry to investigate omeprazole and venlafaxine metabolites in wastewater [J]. Journal of Hazardous Materials, 2016, 302: 332-340. doi: 10.1016/j.jhazmat.2015.09.059 [43] BONNER R, HOPFGARTNER G, SWATH data independent acquisition mass spectrometry for metabolomics [J]. TrAC Trends in Analytical Chemistry, 2018, 120c 115278. [44] BROECKLING C D, HOYES E, RICHARDSON K, et al. Comprehensive tandem-mass-spectrometry coverage of complex samples enabled by data-set-dependent acquisition [J]. Analytical Chemistry, 2018, 90,(13): 8020-8027. doi: 10.1021/acs.analchem.8b00929 [45] PARRILLA VAZQUEZ P, LOZANO A, FERRER C, et al. Improvements in identification and quantitation of pesticide residues in food by LC-QTOF using sequential mass window acquisition (SWATH®) [J]. Analytical Methods, 2018, 10,(24): 2821-2833. doi: 10.1039/C8AY00678D [46] ZHU X, CHEN Y, SUBRAMANIAN R. Comparison of information-dependent acquisition, SWATH, and MS(All) techniques in metabolite identification study employing ultrahigh-performance liquid chromatography-quadrupole time-of-flight mass spectrometry [J]. Analytical Chemistry, 2014, 86(2): 1202-1209. [47] ZHOU J, LI Y, CHEN X, et al. Development of data-independent acquisition workflows for metabolomic analysis on a quadrupole-orbitrap platform [J]. Talanta, 2017, 164: 128-136. doi: 10.1016/j.talanta.2016.11.048 [48] ROEMMELT A T, STEUER A E, KRAEMER T. Liquid chromatography, in combination with a quadrupole time-of-flight instrument, with sequential window acquisition of all theoretical fragment-ion spectra acquisition: validated quantification of 39 antidepressants in whole blood as part of a simultaneous screening and quantification procedure [J]. Analytical Chemistry, 2015, 87(18): 9294-9301. [49] ARMHARD K, GOTTSCHALL A, PITTERL F, et al. Applying 'Sequential Windowed Acquisition of All Theoretical Fragment Ion Mass Spectra' (SWATH) for systematic toxicological analysis with liquid chromatography-high-resolution tandem mass spectrometry [J]. Analytical and Bioanalytical Chemistry, 2015, 407,(2): 405-414. doi: 10.1007/s00216-014-8262-1 [50] ZHOU Z, TU J, XIONG X, SHEN, et al. LipidCCS: Prediction of collision cross-section values for lipids with high precision to support ion mobility-mass spectrometry-based lipidomics [J]. Analytical Chemistry, 2017, 89,(17): 9559-9566. doi: 10.1021/acs.analchem.7b02625 [51] TU J, ZHOU Z, LI T, et al. The emerging role of ion mobility-mass spectrometry in lipidomics to facilitate lipid separation and identification [J]. TrAC Trends in Analytical Chemistry, 2019, 116: 332-339. doi: 10.1016/j.trac.2019.03.017 [52] HELBLING D E, HOLLENDER J, KOHLER H P E, et al. High-throughput identification of microbial transformation products of organic micropollutants [J]. Environmental Science & Technology, 2010, 44,(17): 6621-6627. [53] BERETSOU V G, PSOMA A K, GAGO-FERRERO P, et al. Identification of biotransformation products of citalopram formed in activated sludge [J]. Water Research, 2016, 103: 205-214. doi: 10.1016/j.watres.2016.07.029 [54] MORIYA Y, SHIGEMIZU D, HATTORI M, et al. PathPred: an enzyme-catalyzed metabolic pathway prediction server [J]. Nucleic Acids Resarch, 2010, 38(Web Server issue): W138-143. [55] HUNTSCHA S, HOFSTETTER T B, SCHYMANSHI E L, et al. Biotransformation of benzotriazoles: insights from transformation product identification and compound-specific isotope analysis [J]. Environmental Science & Technology, 2014, 48,(8): 4435-4443. [56] WEIZEL A, SCHLUSENER M P, DIERKES G, et al. Analysis of the aerobic biodegradation of glucocorticoids: Elucidation of the kinetics and transformation reactions [J]. Water Research, 2020: 115561. [57] KRUVE A. Strategies for drawing quantitative conclusions from non-targeted liquid chromatography high-resolution mass spectrometry analysis [J]. Analytical Chemistry, 2020, 92(7): 4691-4699. [58] HUFSKY F, SCHEUBERT K, BOCKER S. New kids on the block: novel informatics methods for natural product discovery [J]. Natural Product Reports, 2014, 31(6): 807-817. [59] CAI W, LI K l, XIONG P, et al. A systematic strategy for rapid identification of chlorogenic acids derivatives in Duhaldea nervosa using UHPLC-Q-Exactive Orbitrap mass spectrometry [J]. Arabian Journal of Chemistry, 2020, 13,(2): 3751-3761. doi: 10.1016/j.arabjc.2020.01.007 [60] QUINN R A, NOTHIAS L F, VINING O, et al. Molecular networking as a drug discovery, drug metabolism, and precision medicine strategy [J]. Trends in Pharmacological Sciences, 2017, 38,(2): 143-154. doi: 10.1016/j.tips.2016.10.011 [61] GAIFFE G, COLE R B, SONNETTE A, et al. Identification of postblast residues by DART-high resolution mass spectrometry combined with multivariate statistical analysis of the Kendrick mass defect [J]. Analytical Chemistry, 2019, 91,(13): 8093-8100. doi: 10.1021/acs.analchem.9b00137 [62] MOREIRA I S, BESSA V S, MURGOLO S, et al. Biodegradation of Diclofenac by the bacterial strain Labrys portucalensis F11 [J]. Ecotoxicology and Environmental Safety, 2018, 152: 104-113. doi: 10.1016/j.ecoenv.2018.01.040 [63] LIU A, SHI J, QU G, et al. Identification of emerging brominated chemicals as the transformation products of tetrabromobisphenol A (TBBPA) derivatives in soil [J]. Environmental Science & Technology, 2017, 51,(10): 5434-5444. [64] WANG Y, YU N, ZHU X, et al. Suspect and nontarget screening of per- and polyfluoroalkyl substances in wastewater from a fluorochemical manufacturing park [J]. Environmental Science & Technology, 2018, 52,(19): 11007-11016. [65] YU N, GUO H, YANG J, et al. Non-target and suspect screening of per- and polyfluoroalkyl substances in airborne particulate matter in China [J]. Environmental Science & Technology, 2018, 52,(15): 8205-8214. [66] SCHOLLEE J E, SCHYMANSKI E L, STRAVS M A, et al. Similarity of high-resolution tandem mass spectrometry spectra of structurally related micropollutants and transformation products [J]. Journal of the American Society for Mass Spectrometry, 2017, 28,(12): 2692-2704. doi: 10.1007/s13361-017-1797-6 [67] YANG J Y, SANCHEZ L M, RATH C M, et al. Molecular networking as a dereplication strategy [J]. Journal of Natural Products, 2013, 76,(9): 1686-1699. doi: 10.1021/np400413s [68] TETA R, DELLA SALA G, GLUKHOV E, et al. Combined LC-MS/MS and molecular networking approach reveals new Cyanotoxins from the 2014 Cyanobacterial Bloom in Green Lake, Seattle [J]. Environmental Science & Technology, 2015, 49,(24): 14301-14310. [69] SHEN X, WANG R, XIONG X, et al. Metabolic reaction network-based recursive metabolite annotation for untargeted metabolomics [J]. Nature Communications, 2019, 10,(1): 1516. doi: 10.1038/s41467-019-09550-x [70] HELBLING D E, HOLLENDER J, KOHLER H P E, et al. Structure-based interpretation of biotransformation pathways of amide-containing compounds in sludge-seeded bioreactors [J]. Environmental Science & Technology, 2010, 44,(17): 6628-6635. [71] HOLLENDER J, SCHYMANSKI E L, SINGER H P, et al. Nontarget screening with high resolution mass spectrometry in the environment: Ready to go? [J]. Environmental Science & Technology, 2017, 51(20): 11505-11512. [72] KIM S, CHEN J, CHENG T, et al. PubChem 2019 update: improved access to chemical data [J]. Nucleic Acids Research, 2019, 47,(D1): D1102-D1109. doi: 10.1093/nar/gky1033 [73] HORAI H, ARITA M, KNAYA S, et al. MassBank: a public repository for sharing mass spectral data for life sciences [J]. Journal of Mass Spectrometry, 2010, 45,(7): 703-714. doi: 10.1002/jms.1777 [74] DOMINGO-ALMENARA X, MONTENEGRO-BURKE J R, IVANISEVIC J, et al. XCMS-MRM and METLIN-MRM: a cloud library and public resource for targeted analysis of small molecules [J]. Nature Methods, 2018, 15,(9): 681-684. doi: 10.1038/s41592-018-0110-3 [75] WOLF S, SCHMIDT S, MULLER-HANNEMANN M, et al. In silico fragmentation for computer assisted identification of metabolite mass spectra [J]. Bmc Bioinformatics, 2010, 11: 12. doi: 10.1186/1471-2105-11-12 [76] SCHYMANSKI E L, GALLMPOIS C M, KRAUSS M, et al. Consensus structure elucidation combining GC/EI-MS, structure generation, and calculated properties [J]. Analytical Chemistry, 2012, 84,(7): 3287-3295. doi: 10.1021/ac203471y [77] SCHYMANSKI E L, JEON J, GULDE R, et al. Identifying small molecules via high resolution mass spectrometry: communicating confidence [J]. Environmental Science & Technology, 2014, 48,(4): 2097-2098. [78] DENG Y, MAO Y, LI B, et al. Aerobic degradation of sulfadiazine by Arthrobacter spp. : Kinetics, pathways, and genomic characterization [J]. Environmental Science & Technology, 2016, 50,(17): 9566-9575. [79] OBEROI A S, JIA Y, ZHANG H, et al. Insights into the fate and removal of antibiotics in engineered biological treatment systems: A critical review [J]. Environmental Science & Technology, 2019, 53,(13): 7234-7264. [80] OOI G T, ESCOLA CASAS M, ANDERSEN H R, et al. Transformation products of clindamycin in moving bed biofilm reactor (MBBR) [J]. Water Research, 2017, 113: 139-148. doi: 10.1016/j.watres.2017.01.058 [81] ACHERMANN S, BIANCO V, MANSFELDT C B, et al. Biotransformation of sulfonamide antibiotics in activated sludge: The formation of pterin-conjugates leads to sustained risk [J]. Environmental Science & Technology, 2018, 52,(11): 6265-6274. [82] ACHERMANN S, FALAS P, JOSS A, et al. Trends in micropollutant biotransformation along a solids retention time gradient [J]. Environmental Science & Technology, 2018, 52,(20): 11601-11611. [83] YU Q, GENG J, HUO H, et al. Bioaugmentated activated sludge degradation of progesterone: Kinetics and mechanism [J]. Chemical Engineering Journal, 2018, 352: 214-224. doi: 10.1016/j.cej.2018.06.159 [84] LIU S S, CHEN J, ZHANG J N, et al. Microbial transformation of progesterone and dydrogesterone by bacteria from swine wastewater: Degradation kinetics and products identification [J]. Science of the Total Environment, 2020, 701: 134930. doi: 10.1016/j.scitotenv.2019.134930 [85] WEIZEL A, SCHLUSENER M P, DIERKES G, et al. Fate and behavior of progestogens in activated sludge treatment: Kinetics and transformation products [J]. Water Research, 2020, 188: 116515. [86] VIENO N, SILLANPAA M. Fate of diclofenac in municipal wastewater treatment plant - a review [J]. Environment International, 2014, 69: 28-39. [87] WU G, GENG J, SHI Y, et al. Comparison of diclofenac transformation in enriched nitrifying sludge and heterotrophic sludge: Transformation rate, pathway, and role exploration [J]. Water Research, 2020, 184: 116158. doi: 10.1016/j.watres.2020.116158 [88] LU Z, SUN W, LI C, et al. Bioremoval of non-steroidal anti-inflammatory drugs by Pseudoxanthomonas sp. DIN-3 isolated from biological activated carbon process [J]. Water Research, 2019, 161: 459-472. doi: 10.1016/j.watres.2019.05.065 [89] BOUJU H, NASTOLD P, BECK B, et al. Elucidation of biotransformation of diclofenac and 4'hydroxydiclofenac during biological wastewater treatment [J]. Journal of Hazardous Materials, 2016, 301: 443-452. doi: 10.1016/j.jhazmat.2015.08.054 [90] JAKIMSKA A, SLIWKA-KASZYNSKA M, RESZCZYNSKA J, et al. Elucidation of transformation pathway of ketoprofen, ibuprofen, and furosemide in surface water and their occurrence in the aqueous environment using UHPLC-QTOF-MS [J]. Analytical and Bioanalytical Chemistry, 2014, 406,(15): 3667-3680. doi: 10.1007/s00216-014-7614-1 [91] OLIVEIRA T S, MURPHY M, MENDOLA N, et al. Characterization of pharmaceuticals and personal care products in hospital effluent and waste water influent/effluent by direct-injection LC-MS-MS [J]. Science of the Total Environment, 2015, 518-519: 459-478. doi: 10.1016/j.scitotenv.2015.02.104 [92] KONIG A, WEIDAUER C, SEIWERT B, et al. Reductive transformation of carbamazepine by abiotic and biotic processes [J]. Water Research, 2016, 101: 272-280. doi: 10.1016/j.watres.2016.05.084 [93] HENNING N, FALAS P, CASTRONOVO S, et al. Biological transformation of fexofenadine and sitagliptin by carrier-attached biomass and suspended sludge from a hybrid moving bed biofilm reactor [J]. Water Research, 2019: 115034. [94] WANG S, WANG J. Biodegradation and metabolic pathway of sulfamethoxazole by a novel strain Acinetobacter sp [J]. Applied Microbiology and Biotechnology, 2018, 102(1): 425-432. [95] SUN F, WU D, CHUA F D, et al. Free nitrous acid (FNA) induced transformation of sulfamethoxazole in the enriched nitrifying culture [J]. Water Research, 2018, 149: 432-439. [96] PENG F Q, YING G G, YANG B, et al. Biotransformation of progesterone and norgestrel by two freshwater microalgae (Scenedesmus obliquus and Chlorella pyrenoidosa): transformation kinetics and products identification [J]. Chemosphere, 2014, 95: 581-588. doi: 10.1016/j.chemosphere.2013.10.013 [97] ARMSTRONG D L, LOZANO N, RICE C P, et al. Degradation of triclosan and triclocarban and formation of transformation products in activated sludge using benchtop bioreactors [J]. Environmental Research, 2018, 161: 17-25. doi: 10.1016/j.envres.2017.10.048 [98] DING T, LIN K, YANG M, et al. Biodegradation of triclosan in diatom Navicula sp. : Kinetics, transformation products, toxicity evaluation and the effects of pH and potassium permanganate [J]. Journal of Hazardous Materials, 2018, 344: 200-209. doi: 10.1016/j.jhazmat.2017.09.033 -



 下载:
下载: