-
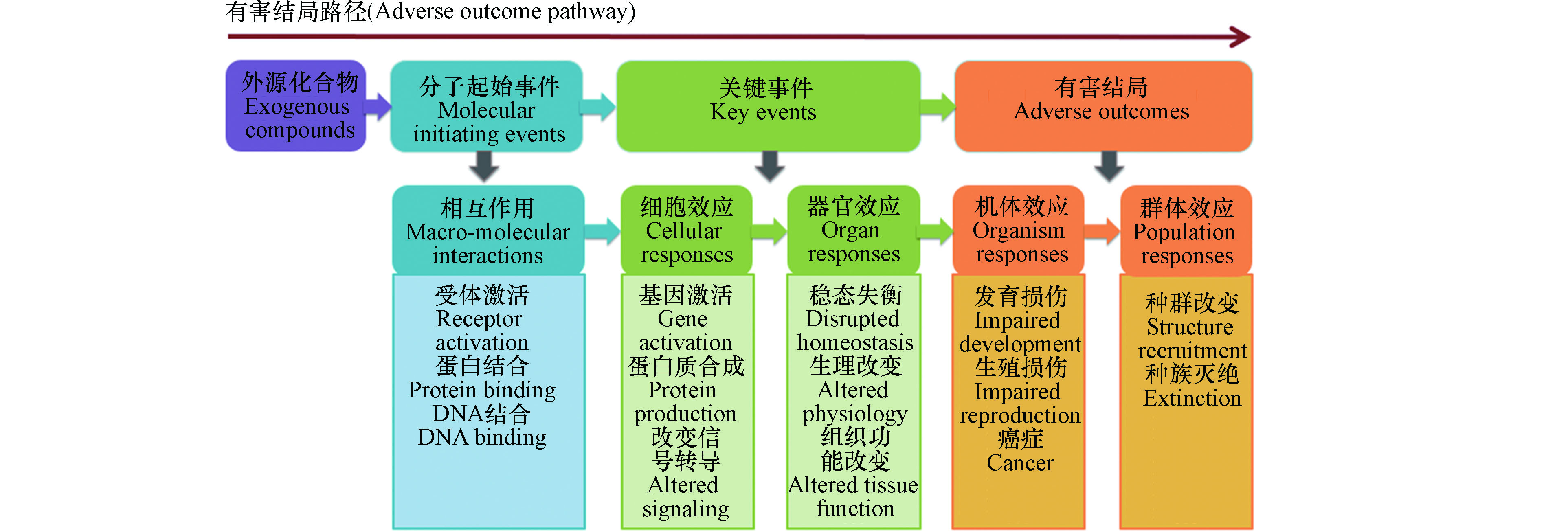
2007年,美国国家研究委员会(National Research Council)发布了具有划时代意义的《21世纪毒性测试:愿景与策略》(Toxicity Testing in the 21st Century: A Vision and a Strategy)报告,指出未来毒性测试和风险评价须考虑毒性通路变化,研究外源化合物暴露导致的基因、蛋白质以及内源性小分子相互作用的生物学通路,以及由此引起的有害健康效应[1-2]。2010年Ankley等进一步提出有害结局路径(adverse outcome pathways, AOPs)这一概念框架[3],主要包括污染物引起的分子起始事件(molecular initiating events, MIEs)以及由其引发的在细胞、组织、器官层次的关键事件(key events, KEs)和最终有害结局,其具体框架如图1所示。AOP基于关键事件关系(key events relationships, KERs),将孤立的KEs有机串联,促进了基于毒性机制的风险评估[4-7]。
MIEs是指外源化合物与毒性通路中特定生物大分子的相互作用,例如外源化合物与DNA、蛋白质分子的共价或非共价作用等[3, 8-10],启动AOP并引起下游KEs,最终导致有害结局。揭示外源化合物MIEs,能够从分子水平明晰外源化合物产生有害健康效应的内在机制,是进行基于毒性作用机制的风险评估的关键。为深入研究外源化合物的毒性作用机制,需要甄别毒性通路中的关键生物大分子,进而考察外源化合物与生物大分子如何相互作用。目前体内、体外实验难以针对大量化合物进行微观尺度实验研究,计算毒理学(Computational Toxicology)方法成为全面化学品风险评估的重要手段,拓宽了MIEs研究的途径。
计算毒理学融合计算化学、化学信息学、毒理学、医学、多组学、系统生物学在内的多学科资源,能够定量评估化学物质典型暴露场景下的暴露特征,对化学物质的理化性质以及毒性进行高通量预测。在原子、分子水平上动态描述外源化合物与生物大分子的相互作用,可助推外源化合物的AOP研究,服务基于毒性作用机制的化学品风险评估与管理[11-14]。《21世纪毒性测试:愿景与策略》、动物福利的“3R原则”(Replacement, Reduction, Refinement)、欧盟REACH法规、美国《劳滕伯格21世纪化学物质安全法案》以及其他相关法规政策,均倡导借助计算毒理学以及体外实验等动物替代方法进行化学品风险评估。今后需更多地借助计算毒理学方法并结合基于人源细胞的体外测试,甄别毒性通路,定量表征MIEs,评估在细胞、组织、个体水平上的关键事件,在分子水平揭示污染物健康危害机制。
关于MIEs研究,随着生物大分子相互作用研究新方法的应用,越来越多研究将理论计算和实验相结合,进行MIEs的甄别、表征、定性及定量研究[15-16],在微观和宏观尺度揭示其相互作用,这有助于深入理解外源化合物如何通过影响生物大分子的生物功能而扰乱相关毒性通路。在相互作用模拟方面,从经典分子动力学模拟,逐渐扩展到粗粒化分子动力学、随机加速动力学、拉伸动力学,以及高斯加速动力学模拟[17-19]。
MIEs研究的难点之一,是甄别关键毒性通路中的生物大分子。随着组学的快速发展[20],通过整合多组学、生物化学、系统生物学以及计算毒理学等方法,可筛选关键毒性通路及大分子受体,进而验证并定量表征MIEs[21]。针对混合物AOP[22],如何综合考虑混合物中各种物质与相同或不同生物大分子的相互作用是MIEs研究的一大挑战,这需进一步甄别混合物各组分如何通过不同MIEs引发相同的有害结局。另外,虽然PDB蛋白质结构数据库(http://www.rcsb.org/)中有大量的生物大分子晶体结构,但与外源化合物相关的复合物晶体结构匮乏,这限制了在原子、分子水平上研究MIEs。DeepMind公司开发的AlphaFold2以及华盛顿大学Davide Baker团队开发的RoseTTAFold,基于深度学习准确预测了蛋白质三维结构[23-24],未来可通过更为精确的计算毒理学方法模建生物大分子的三维结构,为研究污染物与生物大分子的相互作用奠定结构生物学数据基础。
自1956年人工智能诞生以来,机器学习和模式识别已成功用于包括环境健康在内的众多研究,能够针对污染物在细胞和组织水平的有害结局进行建模[25-26]。目前深度学习已成为人工智能研究的主流方向。随着环境大数据的不断增长,基于机器学习、深度学习的计算毒理学在不断发展[27-29]。今后需持续加强污染物毒性数据的收集以及公共数据库的建设与维护,为深度学习准备更为有效的数据,更加高效地服务于基于毒性作用机制的化学品健康风险评估与管理。
分子起始事件在计算毒理学中的研究展望
Perspective of molecular initiating events in computational toxicology
-
摘要: 自2010年有害结局路径(adverse outcome pathways, AOPs)概念框架提出以来,化学品风险评估更加注重围绕关键毒性通路,并强化了分子起始事件(molecular initiating events, MIEs)研究。计算毒理学可在原子、分子水平上研究MIEs,揭示污染物与毒性通路相关的DNA、蛋白质等生物大分子的相互作用,能够促进基于毒性作用机制的化学品风险评估。随着计算毒理学、生物物理学、组学以及人工智能的快速发展,整合计算毒理学以及基于人源细胞的多组学研究,能够快速筛选关键毒性通路,识别相关生物大分子,定量表征MIEs并建立预测模型,强力助推化学品高通量测试及风险评估。Abstract: Since the conceptual framework of adverse outcome pathways (AOPs) was proposed in 2010, chemical risk assessment has been carried out with the emphasis more on key toxicity pathways, and studies on molecular initiating events (MIEs) have been strengthened. Computational toxicology facilitates the investigation of MIEs at the atomic and molecular level, and reveals the interaction between xenobiotics and biomacromolecules such as DNA and proteins, promoting hazard assessment of chemicals. With the rapid development in computational toxicology, biophysics, multi-omics and artificial intelligence, the integration of computational toxicology and human cell-based multi-omics enables rapid screening of key toxicity pathways, identification of biomacromolecules, quantitative characterization of MIEs and the construction of prediction models, significantly enhancing high-throughput testing and risk assessment of chemicals.
-
Key words:
- computational toxicology /
- deep learning /
- interaction /
- adverse outcome pathway /
- risk assessment
-

-
[1] COUNCIL N R. Toxicity testing in the 21st century: a vision and a strategy[R]. Washington DC: The National Academies Press, 2007. [2] GIBB S. Toxicity testing in the 21st century: A vision and a strategy [J]. Reproductive Toxicology (Elmsford, N. Y.), 2008, 25(1): 136-138. doi: 10.1016/j.reprotox.2007.10.013 [3] ANKLEY G T, BENNETT R S, ERICKSON R J, et al. Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment [J]. Environmental Toxicology and Chemistry, 2010, 29: 730-741. doi: 10.1002/etc.34 [4] CONOLLY R B, ANKLEY G T, CHENG W Y, et al. Quantitative adverse outcome pathways and their application to predictive toxicology [J]. Environmental Science & Technology, 2017, 51(8): 4661-4672. [5] KNAPEN D, ANGRISH M M, FORTIN M C, et al. Adverse outcome pathway networks I: Development and applications [J]. Environmental Toxicology and Chemistry, 2018, 37(6): 1723-1733. doi: 10.1002/etc.4125 [6] WITTWEHR C, ALADJOV H, ANKLEY G, et al. How adverse outcome pathways can aid the development and use of computational prediction models for regulatory toxicology [J]. Toxicological Sciences, 2017, 155(2): 326-336. doi: 10.1093/toxsci/kfw207 [7] JEONG J,CHOI J. Use of adverse outcome pathways in chemical toxicity testing: potential advantages and limitations [J]. Environ Health Toxicol, 2017, 33(1): e2018002. doi: 10.5620/eht.e2018002 [8] ALLEN T E H, GOODMAN J M, GUTSELL S, et al. A history of the molecular initiating event [J]. Chemical Research in Toxicology, 2016, 29(12): 2060-2070. doi: 10.1021/acs.chemrestox.6b00341 [9] ALLEN T E H, GOODMAN J M, GUTSELL S, et al. Defining molecular initiating events in the adverse outcome pathway framework for risk assessment [J]. Chemical Research in Toxicology, 2014, 27(12): 2100-2112. doi: 10.1021/tx500345j [10] ELLISON C M, ENOCH S J, CRONIN M T. A review of the use of in silico methods to predict the chemistry of molecular initiating events related to drug toxicity [J]. Expert Opinion on Drug Metabolism & Toxicology, 2011, 7(12): 1481-1495. [11] ELLISON C M, PIECHOTA P, MADDEN J C, et al. Adverse outcome pathway (AOP) informed modeling of aquatic toxicology: QSARs, read-across, and interspecies verification of modes of action [J]. Environmental Science & Technology, 2016, 50(7): 3995-4007. [12] THOMAS R S, BAHADORI T, BUCKLEY T J, et al. The next generation blueprint of computational toxicology at the US environmental protection agency [J]. Toxicological Sciences, 2019, 169(2): 317-332. doi: 10.1093/toxsci/kfz058 [13] KNUDSEN T B, FITZPATRICK S C, de ABREW K N, et al. FutureTox IV workshop summary: Predictive toxicology for healthy children [J]. Toxicological Sciences, 2021, 180(2): 198-211. doi: 10.1093/toxsci/kfab013 [14] 王中钰, 陈景文, 乔显亮, 等. 面向化学品风险评价的计算(预测)毒理学 [J]. 中国科学(化学), 2016, 46(2): 222-240. doi: 10.1360/N032015-00169 WANG Z Y, CHEN J W, QIAO X L, et al. Computational toxicology: Oriented for chemicals risk assessment [J]. Scientia Sinica Chimica, 2016, 46(2): 222-240(in Chinese). doi: 10.1360/N032015-00169
[15] WEDLAKE A J, FOLIA M, PIECHOTA S, et al. Structural alerts and random forest models in a consensus approach for receptor binding molecular initiating events [J]. Chemical Research in Toxicology, 2020, 33(2): 388-401. doi: 10.1021/acs.chemrestox.9b00325 [16] ALLEN T E H, GOODMAN J M, GUTSELL S, et al. Using 2D structural alerts to define chemical categories for molecular initiating events [J]. Toxicological Sciences, 2018, 165(1): 213-223. doi: 10.1093/toxsci/kfy144 [17] MIAO Y L, FEHER V A, MCCAMMON J A. Gaussian accelerated molecular dynamics: Unconstrained enhanced sampling and free energy calculation [J]. Journal of Chemical Theory and Computation, 2015, 11(8): 3584-3595. doi: 10.1021/acs.jctc.5b00436 [18] MIAO Y, MCCAMMON J A. Gaussian accelerated molecular dynamics: theory, implementation, and applications [J]. Annual Reports in Computational Chemistry, 2017, 13: 231-278. [19] ZHAN T J, CUI S X, LIU X J, et al. Enhanced disrupting effect of benzophenone-1 chlorination byproducts to the androgen receptor: cell-based assays and gaussian accelerated molecular dynamics simulations [J]. Chemical Research in Toxicology, 2021, 34(4): 1140-1149. doi: 10.1021/acs.chemrestox.1c00023 [20] BROCKMEIER E K, HODGES G, HUTCHINSON T H, et al. The role of omics in the application of adverse outcome pathways for chemical risk assessment [J]. Toxicological Sciences, 2017, 158(2): 252-262. doi: 10.1093/toxsci/kfx097 [21] SPINU N, CRONIN M T D, ENOCH S J, et al. Quantitative adverse outcome pathway (qAOP) models for toxicity prediction [J]. Arch Toxicol, 2020, 94(5): 1497-1510. doi: 10.1007/s00204-020-02774-7 [22] CHENG F, LI H Z, BROOKS B W, et al. Retrospective risk assessment of chemical mixtures in the big data era: an alternative classification strategy to integrate chemical and toxicological data [J]. Environmental Science & Technology, 2020, 54(10): 5925-5927. [23] JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold [J]. Nature, 2021. DOI: 10.1038/S41586-021-03819-2. [24] BAEK M, DIMAIO F, ANISHCHENKO I, et al. Accurate prediction of protein structures and interactions using a three-track neural network [J]. Science, 2021, 373, 6557: 871-876. DOI: 10.1126/science.abj8754. [25] SCHMIDT C W. Into the black box: what can machine learning offer environmental health research[J]. Environmental Health Perspectives, 2020, 128(2): 22001. [26] LUECHTEFELD T, MARSH D, ROWLANDS C, et al. Machine learning of toxicological big data enables read-across structure activity relationships (RASAR) outperforming animal test reproducibility [J]. Toxicological Sciences, 2018, 165(1): 198-212. doi: 10.1093/toxsci/kfy152 [27] CIALLELLA H, ZHU H. Advancing computational toxicology in the big data era by artificial intelligence: Data-driven and mechanism-driven modeling for chemical toxicity [J]. Chemical Research in Toxicology, 2019, 32(4): 536-547. doi: 10.1021/acs.chemrestox.8b00393 [28] BUTLER K T, DAVIES D W, CARTWRIGHT H, et al. Machine learning for molecular and materials science [J]. Nature, 2018, 559(7715): 547-555. doi: 10.1038/s41586-018-0337-2 [29] ZHU H. Big data and artificial intelligence modeling for drug discovery [J]. Annual Review of Pharmacology and Toxicology, 2020, 60: 573-589. doi: 10.1146/annurev-pharmtox-010919-023324 -



 下载:
下载:

